Bacterial resistance: A major global threat
Antibiotics are indispensable in our battle against bacterial infections. However, the emergence of bacterial resistance is a major threat to global public health and development. A key driver of bacterial resistance is the inappropriate use of antibiotics. Whether through overprescription, misuse, or inappropriate dosing, these practices create selective pressures that favour the evolution of resistant bacterial strains.
Antibiotics in wastewater
Addressing bacterial resistance requires a collective effort that extends beyond the healthcare sector. Current wastewater treatment plants often struggle to effectively remove antibiotics and their transformation products1,2, resulting in the persistence of these active compounds in treated wastewater. Even at low concentrations, these residues contribute to the development of antibiotic resistance, especially when reused for purposes such as irrigation or soil amendment3,4. Physicochemical techniques, such as adsorption5,6, oxidation7 or ozonation8 have shown promise in removing antibiotic residues, but they produce a range of toxic and hazardous byproducts9,10. In response to these challenges, biological techniques have emerged as a sustainable alternative.
Biological alternatives: Fungal biotransformation
Natural processes such as microbial degradation offer a more environmentally friendly approach to antibiotic waste treatment. Biological organisms, particularly fungal and bacterial species, produce extracellular ligninolytic enzymes, such as laccases and haem-peroxidases, which catalyse the transformation of antibiotics11. These biotransformation processes follow different pathways depending on the organism involved and the structure of the antibiotic.
Biotransformation of levofloxacin by basidiomycete fungi
A recent study by several researchers from Tunisia, France and the USA set out to investigate the potential of two basidiomycete fungal strains to transform levofloxacin-contaminated wastewater12. Levofloxacin is an antibiotic in the fluoroquinolone group used to treat infections of the eye, bladder and severe lung infections. When taken orally, levofloxacin is almost completely absorbed from the intestine into the bloodstream and is mainly excreted unchanged by the kidneys and therefore ends up in our wastewater.
The fungi Porostereum spadiceum and Bjerkandera adusta were selected for their demonstrated ability to degrade a range of challenging micropollutants, such as dyes, phenols, and other pharmaceuticals13,14. During a 10-day culture period, both fungi were evaluated for their ability to biotransform levofloxacin. P. spadiceum completely degraded levofloxacin by day 10, making it the preferred strain for further experiments. The researchers sequenced its genome, provided insights into the potential enzymes involved in the biotransformation process, and analysed the products of the biotransformed levofloxacin using high-resolution mass spectrometry.
Mechanistic insights: Enzymatic activity
The study investigated the potential involvement of oxidative enzymes, focusing on the extracellular enzymes involved in lignin degradation, i.e. class II peroxidases and laccases. No laccase-like activity was detected. Peroxidase-like activity peaked at day 8, indicating that haem-peroxidases are the primary enzymes produced by P. spadiceum. As there was no significant difference for these enzymes in the presence or absence of levofloxacin, the enzyme machinery appears to be neither induced nor repressed by levofloxacin. To further elucidate the enzymes responsible for levofloxacin degradation, the genome of P. spadiceum was sequenced, and its main carbohydrate-active enzymes were annotated. Predictions suggested the presence of lignin-degrading activities potentially involved in the biotransformation of levofloxacin. The presence of levofloxacin in the medium induced the overexpression of aryl alcohol oxidase and glucose 1-oxidase. These enzymes play a crucial role in providing hydrogen peroxide (H2O2), a necessary component for the oxidation of aromatic compounds and lignin, which supports the activity of haem-peroxidases and other enzymes involved in the modification of levofloxacin15.
Analysis of biotransformation products using SIRIUS
Aiming to identify levofloxacin degradation products, the researchers used ultra-high performance liquid chromatography coupled to high-resolution mass spectrometry to analyze secretome extracts of P. spadiceum. Spectra were annotated individually and corroborated with SIRIUS and CSI:FingerID annotations. Levofloxacin was still detectable on UV chromatograms on days 4 and 7, but was absent by day 10. No degradation products were detected on day 4, but five major degradation products emerged in UV chromatograms thereafter. All major biotransformation processes appeared to occur mainly at the piperazine ring (oxidation reactions and ring cleavage). Unfortunately, spectral comparison with purified reference standards is not possible at present as they are not commercially available.
Compound 1 was detected only on day 10 and is a possible transformation from the most abundant product remaining (i.e. compound 4) replacing the primary amine with a hydroxyl group.
Compoud 2 may be produced from compound 3 through the loss of the aliphatic chain in the alpha position of the amide function.
The fragmentation spectrum of compound 3 indicated oxydation and cleavage of the piperazine cycle and three fragments were formed by successive loss of the carbonyl function.
Compound 4 is levofloxacin with a primary amine in place of the piperazine cycle, as previously described for ofloxacin16.
Compound 5 was detected only at trace levels on days 7 and 10 and likely appeared due to the loss of the carbonyl function from the tertiary amine of compound 3.
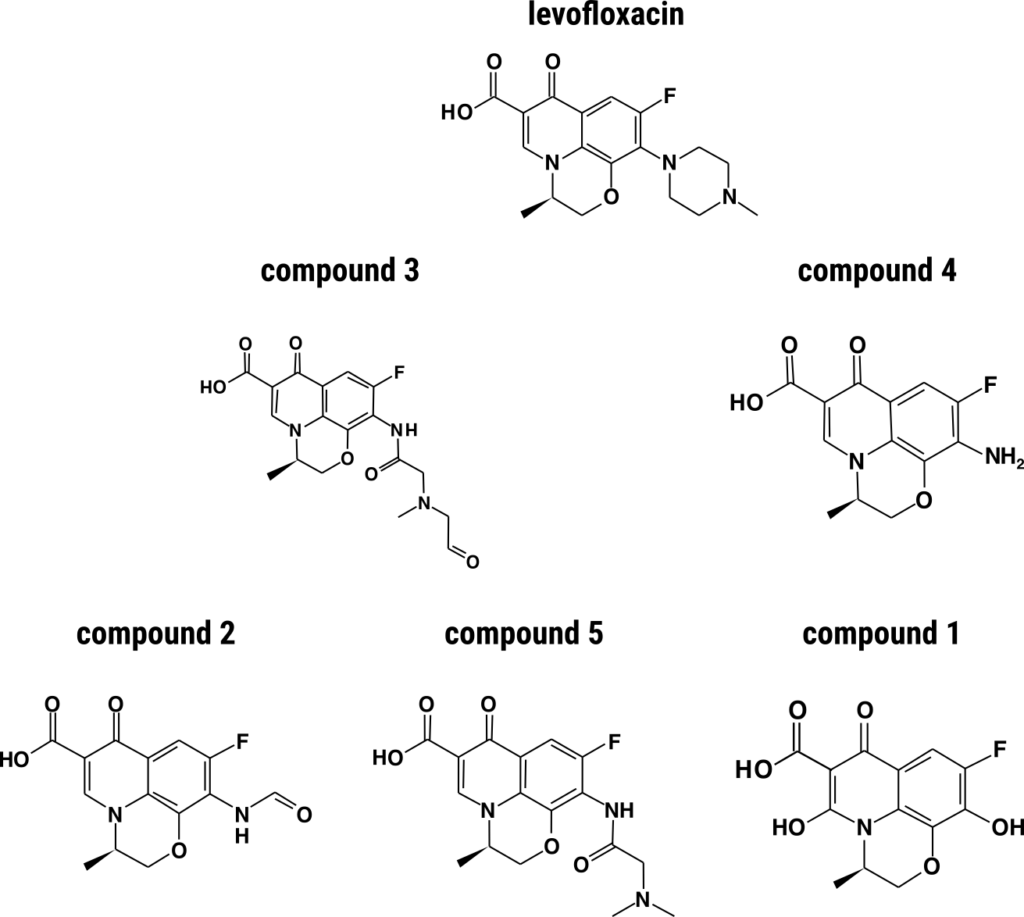
Unfortunately, the five major degradation products of levofloxacin retained their quinolone core, and thus may retain some of their antibiotic properties, especially against Gram-positive bacteria17. To achieve biotransformation of the quinolone antibiotic core, co-culturing of different basidiomycete strains known for their complementary metabolic properties should be investigated in the future.
References
- 1.Phoon BL, Ong CC, Mohamed Saheed MS, et al. Conventional and emerging technologies for removal of antibiotics from wastewater. Journal of Hazardous Materials. Published online December 2020:122961. doi:10.1016/j.jhazmat.2020.122961
- 2.Langbehn RK, Michels C, Soares HM. Antibiotics in wastewater: From its occurrence to the biological removal by environmentally conscious technologies. Environmental Pollution. Published online April 2021:116603. doi:10.1016/j.envpol.2021.116603
- 3.Oberoi AS, Jia Y, Zhang H, Khanal SK, Lu H. Insights into the Fate and Removal of Antibiotics in Engineered Biological Treatment Systems: A Critical Review. Environ Sci Technol. Published online June 3, 2019:7234-7264. doi:10.1021/acs.est.9b01131
- 4.Wang D, Ning Q, Dong J, Brooks BW, You J. Predicting mixture toxicity and antibiotic resistance of fluoroquinolones and their photodegradation products in Escherichia coli. Environmental Pollution. Published online July 2020:114275. doi:10.1016/j.envpol.2020.114275
- 5.Gao B, Chang Q, Xi Z, El-Sayed MMH, Shoeib T, Yang H. Fabrication of environmentally-friendly composited sponges for efficient removal of fluoroquinolones antibiotics from water. Journal of Hazardous Materials. Published online March 2022:127796. doi:10.1016/j.jhazmat.2021.127796
- 6.Wu Q, Zhang Y, Cui M hua, et al. Pyrolyzing pharmaceutical sludge to biochar as an efficient adsorbent for deep removal of fluoroquinolone antibiotics from pharmaceutical wastewater: Performance and mechanism. Journal of Hazardous Materials. Published online March 2022:127798. doi:10.1016/j.jhazmat.2021.127798
- 7.Kim J, Coulibaly GN, Yoon S, Assadi AA, Hanna K, Bae S. Red mud-activated peroxymonosulfate process for the removal of fluoroquinolones in hospital wastewater. Water Research. Published online October 2020:116171. doi:10.1016/j.watres.2020.116171
- 8.González-Labrada K, Richard R, Andriantsiferana C, Valdés H, Jáuregui-Haza UJ, Manero MH. Enhancement of ciprofloxacin degradation in aqueous system by heterogeneous catalytic ozonation. Environ Sci Pollut Res. Published online November 27, 2018:1246-1255. doi:10.1007/s11356-018-3559-9
- 9.Crini G, Lichtfouse E. Advantages and disadvantages of techniques used for wastewater treatment. Environ Chem Lett. Published online July 31, 2018:145-155. doi:10.1007/s10311-018-0785-9
- 10.Bhatt S, Chatterjee S. Fluoroquinolone antibiotics: Occurrence, mode of action, resistance, environmental detection, and remediation – A comprehensive review. Environmental Pollution. Published online December 2022:120440. doi:10.1016/j.envpol.2022.120440
- 11.Manasfi R, Chiron S, Montemurro N, Perez S, Brienza M. Biodegradation of fluoroquinolone antibiotics and the climbazole fungicide by Trichoderma species. Environ Sci Pollut Res. Published online April 27, 2020:23331-23341. doi:10.1007/s11356-020-08442-8
- 12.Ben Ayed A, Akrout I, Staita K, et al. Genome sequencing of Porostereum spadiceum to study the degradation of levofloxacin. Ecotoxicology and Environmental Safety. Published online January 2024:115808. doi:10.1016/j.ecoenv.2023.115808
- 13.Gao T, Qin D, Zuo S, et al. Decolorization and detoxification of triphenylmethane dyes by isolated endophytic fungus, Bjerkandera adusta SWUSI4 under non-nutritive conditions. Bioresour Bioprocess. Published online October 6, 2020. doi:10.1186/s40643-020-00340-8
- 14.Dhiman N, Chaudhary S, Singh A, Chauhan A, Kumar R. Sustainable degradation of pharmaceutical waste using different fungal strains: Enzyme induction, kinetics and isotherm studies. Environmental Technology & Innovation. Published online February 2022:102156. doi:10.1016/j.eti.2021.102156
- 15.Sützl L, Laurent CVFP, Abrera AT, Schütz G, Ludwig R, Haltrich D. Multiplicity of enzymatic functions in the CAZy AA3 family. Appl Microbiol Biotechnol. Published online February 6, 2018:2477-2492. doi:10.1007/s00253-018-8784-0
- 16.Jimenez-Villarin J, Serra-Clusellas A, Martínez C, Conesa A, Garcia-Montaño J, Moyano E. Liquid chromatography coupled to tandem and high resolution mass spectrometry for the characterisation of ofloxacin transformation products after titanium dioxide photocatalysis. Journal of Chromatography A. Published online April 2016:201-210. doi:10.1016/j.chroma.2016.03.063
- 17.Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics. Med Chem Commun. Published online 2019:1719-1739. doi:10.1039/c9md00120d








