Why Should We Care About Pharmaceutical Contamination?
Pharmaceutical pollution in water is a growing issue, posing potential risks to ecosystems and human health. Active pharmaceutical ingredients can disrupt ecosystems, harm aquatic life, and affect biodiversity. The presence of antibiotics in the environment contributes to the development of drug-resistant bacteria. And the effects of long-term exposure to these contaminants in drinking water and food chains on human health are unknown.
Transformation Products – the Hidden Threads
While traditional environmental monitoring methods often focus on known drug compounds, their transformation products—chemical derivatives formed as drugs break down—can be more persistent or bioaccumulative with higher bioactivity than the original drug. Moreover, transformation products may escape conventional wastewater treatment processes. Non-targeted screening using high-resolution mass spectrometry allows detection of both known and unknown contaminants. Expanding analytical strategies to include transformation products enables a deeper understanding of their environmental fate and behavior, which is essential for developing effective mitigation strategies to safeguard public health.
Investigating Pharmaceuticals in Luxembourg’s Waterways
We annotate pharmaceuticals in Luxembourgish rivers using precursor drug screening and transformation product screening with SIRIUS. We use data obtained by a research team around Emma Schymanski from University of Luxembourg1. They collected 92 surface water samples from Luxembourgish rivers at 13 different locations during routine monitoring events between 2019 and 2020 and analyzed them with liquid chromatography coupled to a high-resolution mass spectrometer (LC-HRMS).
We analysed the data relying on different resources.
- Pharmaceutical Drug List: 816 approved drugs in Luxembourg.
- Spectral Libraries: MassBank and MoNA, which contain reference spectra for 577 of the listed drugs.
- Custom Drug Database: Molecular structures for the listed drugs. This database contains 772 molecular structures. For stereoisomers, one representative is used; compounds with invalid SMILES notations were discarded.
- Transformation Product Database: Generated using BioTransformer2,3, this database contains over 1.06 million potential transformation products for 713 of the listed drugs, ultimately filtered down to 483,203 unique structures.
How to Screen for Pharmaceuticals in an Environmental Sample
All 92 water samples yielded 15,819 valid features after preprocessing. This includes features of all quality levels, but no features containing only MS1 spectra, being multiply charged, or being multimeric. SIRIUS assigned molecular formulas based on the listed databases. The structure annotation workflow in SIRIUS is a non-targeted workflow. SIRIUS identifies the structure of a molecule by predicting its molecular fingerprint and searching for matches in a molecular structure database4. Here, we demonstrate how SIRIUS detects precursor drugs as well as transformation products using custom databases. We search within biomolecule structure databases (part of SIRIUS) as well as the imported Custom Drug Database and Transformation Product Database. For validation, we perform spectral library searches in MassBank and MoNA. Since we have not experimentally verified the results, all identifications remain putative. For more details on how to conduct transformation product screening in SIRIUS, read this blog post.

Finding Precursor Drugs
SIRIUS reported 80 drugs from the drug list as top hits (right, pink). Among these, 30 could not be matched by spectral library search. From these drugs that would not have been reported by spectral library search, 17 were confidently annotated by SIRIUS5 (that means they received a COSMIC confidence score of more than 0.64, which roughly corresponds to FDR 10 % in evaluations), proving its ability to detect pharmaceuticals that traditional methods might overlook. 50 compounds have been identified by both methods.

Preventing Suspect Blindness
SIRIUS offers the possibility to investigate deeper into the results to explore the structural explanations. In the Structures view, you can see how well a structure is supported by fingerprint evidence. The Substructure Annotation view, helps you investigate the direct connection between the input MS/MS spectrum and the structure candidates.
In our example, one feature needs deeper investigation. Spectral matching suggested this feature to be aspirin (acetylsalicylic acid), while the best SIRIUS match for this feature was monomethyl phthalate, which had an equally high spectral library search score (both cosine similarities ∼0.97) but is not included in the drug list. Monomethyl phthalate is a breakdown product of dimethyl phthalate, used in insect repellant and plastic, justifying the possibility of this alternative putative identification. Further investigation into both results could provide additional clarification. The unambiguous identity of this feature can only be resolved by additional experimental data. The advantage of using SIRIUS lies in its ability to present both possibilities—Aspirin as the best spectral match and monomethyl phthalate as the best structure database match—preventing ”suspect-blindness” and showing that the annotation of Aspirin is not unambiguous.
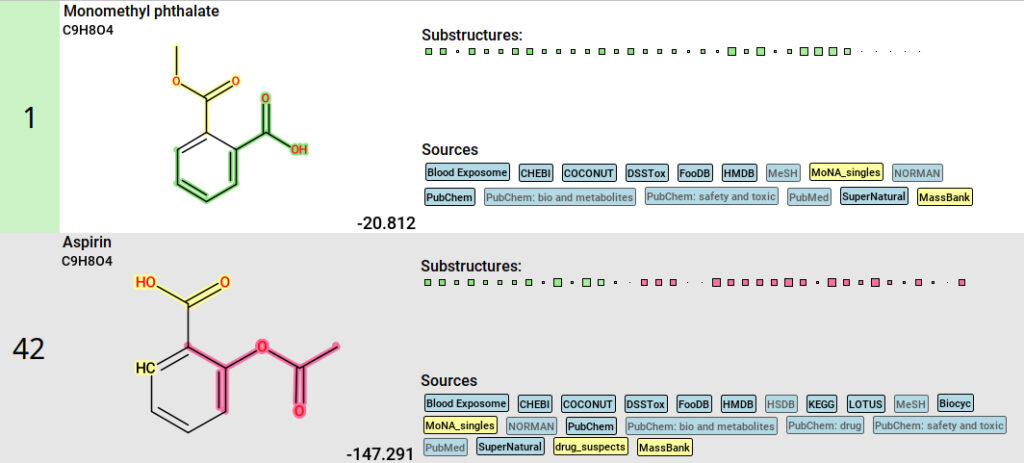
SIRIUS results for monomethyl phthalate and Aspirin. The color highlighting of the candidate structures reflects substructures that are supported by fingerprint evidence (green), substructures that contradict fingerprint evidence (pink), and substructures with mixed support (yellow).
Uncovering Hidden Transformation Products
When screening for transformation products not already on the drug list, we detected 292 unique transformation products (pink). Only 33 of these were found using spectral library search (blue), proving that most would have gone undetected relying on spectral library search only. 85 transformation products were classified as high-confidence identifications, with 56 of these completely absent from spectral libraries. These results emphasize how advanced screening methods can reveal hidden pollutants that may pose environmental and health risks.

Beyond Pharmaceuticals
Screening for transformation products is not just important for pharmaceuticals—it can also be applied to pesticides and industrial chemicals, which break down into potentially hazardous transformation products. SIRIUS enhances environmental monitoring by searching in transformation product databases, and integrating multiple validation levels, including spectral library searches, confidence scores for database hits, and substructure annotations for deeper analysis.

A New Era of Environmental Monitoring
SIRIUS offers a cutting-edge approach to tracking pharmaceutical contamination. By identifying both precursor drugs and transformation products, it provides a clearer picture of the pollutants present in our waterways. The ability to detect transformation products without requiring reference spectra makes SIRIUS invaluable for discovering emerging contaminants.
Want to learn more?
Download the full application note (first published in Int. Labmate 50.3 April 2025) or read the “How to” on transformation product screening.
References
- Randolph R. Singh et al. “Occurrence and Distribution of Pharmaceuticals and Their Transformation Products in Luxembourgish Surface Waters”. In: ACS Environmental Au 1.1 (July 2021), pp. 58–70. ISSN: 2694-2518. DOI: 10.1021/acsenvironau.1c00008.
- Yannick Djoumbou-Feunang et al. “BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification”. In: J. Cheminf. 11.1 (Jan. 2019). ISSN: 1758-2946. DOI: 10.1186/s13321-018-0324-5.
- David S Wishart et al. “BioTransformer 3.0—a web server for accurately predicting metabolic transformation products”. In: Nucleic Acids Res. 50.W1 (May 2022), W115–W123. ISSN: 1362-4962. DOI: 10.1093/nar/gkac313.
- Kai Dührkop et al. “Searching molecular structure databases with tandem mass spectra using CSI:FingerID”. In: Proc. Natl. Acad. Sci. 112.41 (Sept. 2015), pp. 12580–12585. ISSN: 1091-6490. DOI: 10.1073/pnas.1509788112.
- Martin A. Hoffmann et al. “High-confidence structural annotation of metabolites absent from spectral libraries”. In: Nat. Biotechnol. 40.3 (Oct. 2021), pp. 411–421. ISSN: 1546-1696. DOI: 10.1038/s41587-021-01045-9.





