Thawing permafrost: the hidden threat
Climate change is a pressing global issue with far-reaching consequences, the extent of which we are still far from understanding. One such effect is the thawing of permafrost. Permafrost is a layer of soil that has remained frozen for at least two consecutive years and is found in high latitude regions, such as the Arctic. It acts as a massive carbon sink, storing an estimated 50% of the world’s soil carbon1,2, which is almost double the amount of carbon in the atmosphere3. The effects of thawing permafrost are far-reaching, affecting the Arctic ecosystems as well as the global climate. As global temperatures rise, permafrost begins to thaw, releasing stored carbon that is decomposed by resident microbial communities into carbon dioxide and methane; potent greenhouse gases that further accelerate global warming4,5.
Enzyme latch hypothesis: Polyphenols controlling the carbon cycle in peatlands
The enzyme latch hypothesis suggests that when there is insufficient oxygen in wetlands, the enzymes that break down organic matter are slowed down and thus carbon is stored. Polyphenols are a chemically diverse and abundant group6 which are decomposed by oxygen-requiring phenol oxidases7,8. Saturated soil conditions following permafrost thaw would render phenol oxidases inactive and ultimately stop microbial carbon decomposition to stabilise soil carbon. Several studies have found conflicting support for parts of the enzyme latch hypothesis9–12 and the possibility of alternative microbial metabolic strategies is so far unexplored.
Beyond phenol oxidases
There is some evidence against phenol oxidases being the only enzymes that degrade polyphenols. Plants have been producing polyphenols for hundreds of millions of years13,14, and microbes that are frequently exposed to them have probably evolved different mechanisms to utilise or resist them15. Moreover, in the human gut other microbial enzymes have been discovered that degrade various polyphenols even in the absence of oxygen16.
New insights from Stordalen Mire
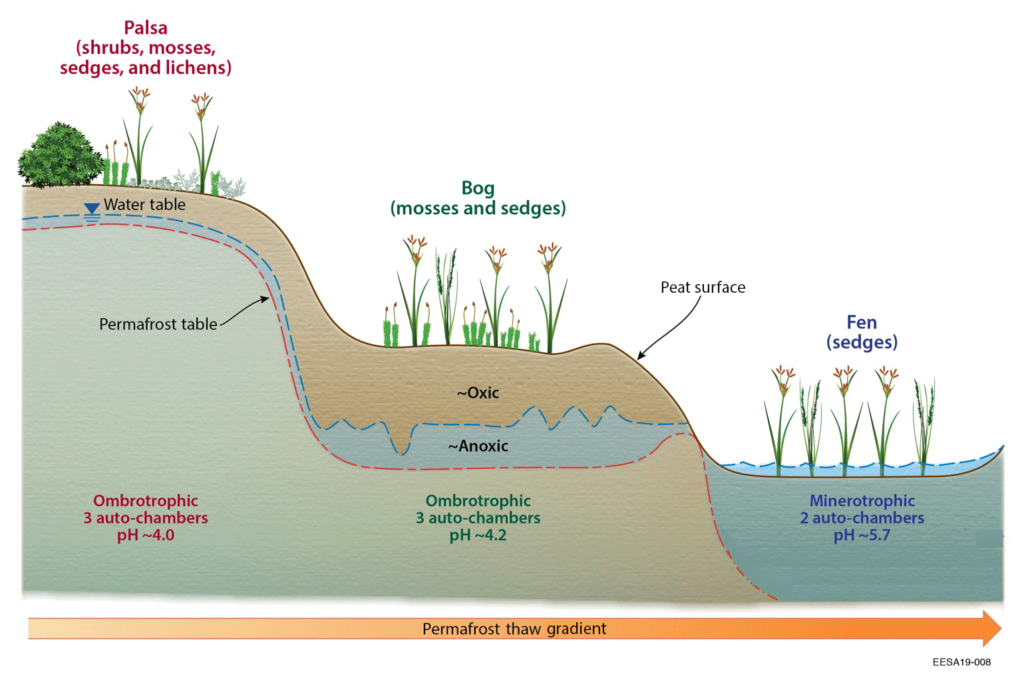
A group of researchers from the USA and Australia led by Kelly C. Wrighton at Colorado State University, USA, have revisited the basic ideas of the enzyme latch hypothesis and investigated how microbes in peatland soils transform polyphenols17. Stordalen Mire is an Arctic peatland, where natural thaw has created three different habitats: intact permafrost palsa, partially thawed bog, and fully thawed fen (see figure). The research team obtained paired genome-resolved metatranscriptome, metabolite and geochemical data to provide a new perspective on polyphenols in the microbially catalysed carbon cycle in thawing permafrost.
For the metabolomic data, water-soluble metabolites were extracted from peat and analysed by untargeted UHPLC-MS/MS. Metabolite annotation was initially performed using an internal database and extended using SIRIUS18, CSI:FingerID19 and CANOPUS20.
Seamless Integration of Spectral Library Search
In SIRIUS 6 we offer smooth integration of your long-established annotation workflow of in-house suspect libraries with comprehensive structure annotation by SIRIUS. No more tedious manual merging – we’ve streamlined the process to enhance your workflow. Identification results will be ranked by CSI:FingerID score and spectral library hits will be added to provide you with a comprehensive view of your data.
No more digging through separate result lists – it’s all in one place!
The talented microbiome
Summarising the extensive results of the study, it is clear above all that the enzyme latch theory is not a suitable description for the dynamics of carbon processing in the Stordalen Mire. The microbial community has adapted its gene expression strategies to the habitat and the oxygen conditions prevailing there. Phenol oxidases alone do not control the fate of polyphenols and the presence or absence of oxygen is not an on/off switch for polyphenol degradation. Rather, polyphenols are integrated into a larger carbon degradation network, including a variety of polyphenol metabolic processes. The decomposition of polyphenols into smaller phenols can even boost carbon dioxide and carbon monoxide production.
Assessing the consequences
The biggest question arising from this study is how these findings will affect carbon storage. We cannot assume that carbon in thawing permafrost will remain stored due to a lack of oxygen. Addressing the challenges posed by thawing permafrost requires a concerted effort to mitigate climate change. While the situation is complex, understanding the dynamics of permafrost and its relationship with climate change is essential for developing strategies to cope with its effects.
References
- 1.Hugelius G, Strauss J, Zubrzycki S, et al. Estimated stocks of circumpolar permafrost carbon with quantified uncertainty ranges and identified data gaps. Biogeosciences. Published online December 1, 2014:6573-6593. doi:10.5194/bg-11-6573-2014
- 2.Schuur EAG, Abbott BW, Commane R, et al. Permafrost and Climate Change: Carbon Cycle Feedbacks From the Warming Arctic. Annu Rev Environ Resour. Published online October 17, 2022:343-371. doi:10.1146/annurev-environ-012220-011847
- 3.Natali SM, Holdren JP, Rogers BM, et al. Permafrost carbon feedbacks threaten global climate goals. Proc Natl Acad Sci USA. Published online May 17, 2021. doi:10.1073/pnas.2100163118
- 4.Woodcroft BJ, Singleton CM, Boyd JA, et al. Genome-centric view of carbon processing in thawing permafrost. Nature. Published online July 16, 2018:49-54. doi:10.1038/s41586-018-0338-1
- 5.Koven CD, Ringeval B, Friedlingstein P, et al. Permafrost carbon-climate feedbacks accelerate global warming. Proc Natl Acad Sci USA. Published online August 18, 2011:14769-14774. doi:10.1073/pnas.1103910108
- 6.Zak D, Roth C, Unger V, et al. Unraveling the Importance of Polyphenols for Microbial Carbon Mineralization in Rewetted Riparian Peatlands. Front Environ Sci. Published online October 4, 2019. doi:10.3389/fenvs.2019.00147
- 7.Freeman C, Ostle N, Kang H. An enzymic “latch” on a global carbon store. Nature. Published online January 2001:149-149. doi:10.1038/35051650
- 8.Fenner N, Freeman C. Woody litter protects peat carbon stocks during drought. Nat Clim Chang. Published online March 16, 2020:363-369. doi:10.1038/s41558-020-0727-y
- 9.Urbanová Z, Hájek T. Revisiting the concept of ‘enzymic latch’ on carbon in peatlands. Science of The Total Environment. Published online July 2021:146384. doi:10.1016/j.scitotenv.2021.146384
- 10.Hall SJ, Treffkorn J, Silver WL. Breaking the enzymatic latch: impacts of reducing conditions on hydrolytic enzyme activity in tropical forest soils. Ecology. Published online October 2014:2964-2973. doi:10.1890/13-2151.1
- 11.Brouns K, Keuskamp JA, Potkamp G, Verhoeven JTA, Hefting MM. Peat origin and land use effects on microbial activity, respiration dynamics and exo-enzyme activities in drained peat soils in the Netherlands. Soil Biology and Biochemistry. Published online April 2016:144-155. doi:10.1016/j.soilbio.2015.11.018
- 12.Romanowicz KJ, Kane ES, Potvin LR, Daniels AL, Kolka RK, Lilleskov EA. Understanding drivers of peatland extracellular enzyme activity in the PEATcosm experiment: mixed evidence for enzymic latch hypothesis. Plant Soil. Published online December 2015:371-386. doi:10.1007/s11104-015-2746-4
- 13.Renault H, Alber A, Horst NA, et al. A phenol-enriched cuticle is ancestral to lignin evolution in land plants. Nat Commun. Published online March 8, 2017. doi:10.1038/ncomms14713
- 14.Berland H, Albert NW, Stavland A, et al. Auronidins are a previously unreported class of flavonoid pigments that challenges when anthocyanin biosynthesis evolved in plants. Proc Natl Acad Sci USA. Published online September 16, 2019:20232-20239. doi:10.1073/pnas.1912741116
- 15.Scalbert A. Antimicrobial properties of tannins. Phytochemistry. Published online January 1991:3875-3883. doi:10.1016/0031-9422(91)83426-l
- 16.Goris T, Cuadrat RRC, Braune A. Flavonoid-Modifying Capabilities of the Human Gut Microbiome—An In Silico Study. Nutrients. Published online August 3, 2021:2688. doi:10.3390/nu13082688
- 17.McGivern BB, Cronin DR, Ellenbogen JB, et al. Microbial polyphenol metabolism is part of the thawing permafrost carbon cycle. Nat Microbiol. Published online May 28, 2024:1454-1466. doi:10.1038/s41564-024-01691-0
- 18.Dührkop K, Fleischauer M, Ludwig M, et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. Published online March 18, 2019:299-302. doi:10.1038/s41592-019-0344-8
- 19.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. Published online September 21, 2015:12580-12585. doi:10.1073/pnas.1509788112
- 20.Dührkop K, Nothias LF, Fleischauer M, et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat Biotechnol. Published online November 23, 2020:462-471. doi:10.1038/s41587-020-0740-8
- 21.Chang KY, Riley WJ, Crill PM, Grant RF, Rich VI, Saleska SR. Large carbon cycle sensitivities to climate across a permafrost thaw gradient in subarctic Sweden. The Cryosphere. Published online February 22, 2019:647-663. doi:10.5194/tc-13-647-2019








