Opportunistic infections in cystic fibrosis
Cystic fibrosis is an inherited disorder that causes severe damage to the lungs, digestive system, and other organs. Opportunistic pathogens like Pseudomonas aeruginosa and Staphylococcus aureus exploit weaknesses in our defense systems to establish infections. Both bacteria are among the most common coinfecting bacteria in human infections, including the cystic fibrosis (CF) lung1,2. Coinfection with these microbes enhances disease severity and antimicrobial tolerance3.
Befriend your competitor: dual-species biofilms
In certain cocultures (e.g. in plankton), the presence of P. aeruginosa reduces the viability of S. aureus4. However, under different conditions, S. aureus can not only survive the presence of P. aeruginosa but the two species can actually thrive together in a dual-species biofilm. Biofilms are highly organized structures that nurture bacteria and protect them from external factors such as the host’s immune system and antibiotic treatments5,6. The cooperation between P. aeruginosa and S. aureus in cystic fibrosis leads to persistent colonization of human airways, increased disease severity and antibiotic resistance.
Quorum sensing: communicate with your neighbour
So far, only little is known about the chemical communication mechanisms between competing bacteria in multi-species environments that enable both species to adapt and survive, and perhaps even thrive. Quorum sensing is a process of cell–cell communication that allows bacteria to share information about population density through the secretion of signaling molecules and adjust gene expression accordingly7. The secretion of those chemical signals depends on the growth status and environmental conditions surrounding the bacteria. The metabolic profile of bacteria in planktonic cultures, for example, differs significantly from their metabolic profile in biofilms.
Metabolic profiles in mutual biofilms
In a recent study, researchers from the Ben Gurion University of the Negev, Israel, used untargeted mass spectrometry-based metabolomics analyses to identify specific molecules that mediate interactions between P. aeruginosa and S. aureus in dual-species biofilms8. They employed high-resolution tandem mass spectrometry coupled with ultrahigh-performance liquid chromatography (UHPLC-HRMS/MS) to detect and characterize these signaling molecules that may regulate chemical communication between bacteria.
The researchers found that the relative inoculum size of both species plays a crucial role in the establishment of a successful mutual biofilm. The metabolic profile of the mixed samples resembles P. aeruginosa monocultures more than S. aureus monocultures. But with increased concentration of S. aureus, the metabolic profile of the biofilm drifts away from that of P. aeruginosa. They observed specific changes in P. aeruginosa metabolite secretion in response to the presence of S. aureus.
Competing for iron
The researchers investigated the molecules secreted by P. aeruginosa, whose abundance depends on S. aureus population density. Several relevant metabolites were identified in GNPS9. One particular metabolite whose increase in concentration is correlated to S. aureus abundance is pyochelin methyl ester (PME). PME is a derivative of pyochelin, a siderophore secreted by P. aeruginosa. Siderophores scavenge iron from the surroundings and import it into the bacterial cells10,11. P. aeruginosa and S. aureus compete for iron, an essential nutrient for bacterial growth. The researchers discovered that pyochelin consistently decreases as the content of S. aureus in the mixed biofilm increases, while the concentration of PME increases. S. aureus possesses an enzyme that converts pyochelin into PME12, which reduces its affinity for iron. This conversion disables P. aeruginosa‘s ability to effectively scavenge iron and may elevate the fitness of S. aureus in the dual-species biofilm. But PME may not only protect S. aureus; it could also serve as a signaling molecule for P. aeruginosa. By recognizing PME, P. aeruginosa may be able to sense the presence of S. aureus and modify its behavior accordingly. This could involve adjusting its virulence toward the potential competitor and either competing or cooperating, depending on the relative population densities of the two species.
Not only a by-product
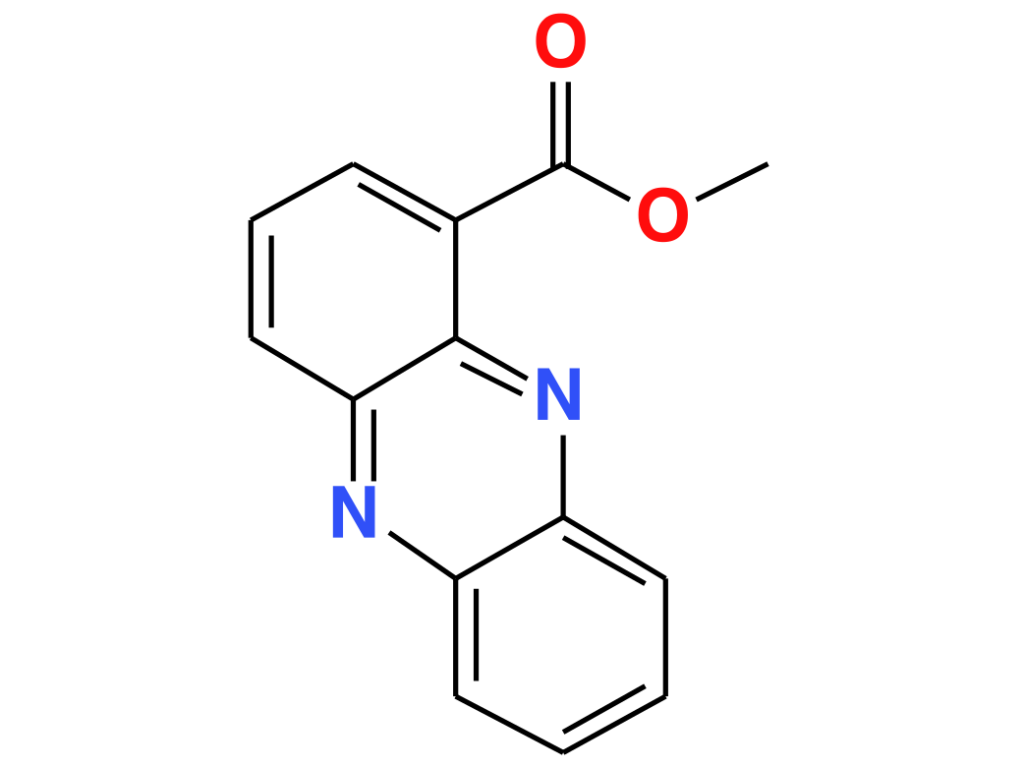
PME seems to be more than a by-product of P. aeruginosa – S. aureus encounters. The researchers investigated the influence of PME on P. aeruginosa‘s metabolism and found that it induced the production of a certain metabolite. This metabolite does not appear in the GNPS database, so they used SIRIUS and CSI:FIngerID13,14 to gain more information on its structure. CSI:FingerID annotates this metabolite as methyl phenazine-1-carboxylate (MPCA), a derivative of phenazine-1-carboxylic acid (PCA), with 91% certainty. The annotation was confirmed through preparation of the respective standard and comparison of NMR and UPLC-HRMS data (level I structure identification15). Phenazines play a role in virulence and antimicrobial properties16,17. Some phenazines, such as PCA, are used by P. aeruginosa for iron acquisition under limiting conditions18,19. MPCA was previously found in soil samples containing Streptomyces canus20, and showed antifungal activities, stronger than PCA in some cases21,22. It was also tested for antibacterial activity against MRSA but exhibited only mild inhibitory effects23. The influence of PME on phenazine production could be related to the increased pathogenesis of this dual-species biofilm5,6.
References
- 1.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance—the need for global solutions. The Lancet Infectious Diseases. Published online December 2013:1057-1098. doi:10.1016/s1473-3099(13)70318-9
- 2.Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Research International. Published online 2016:1-8. doi:10.1155/2016/2475067
- 3.Barraza JP, Whiteley M. A Pseudomonas aeruginosa Antimicrobial Affects the Biogeography but Not Fitness of Staphylococcus aureus during Coculture. Limoli DH, Gilmore MS, eds. mBio. Published online April 27, 2021. doi:10.1128/mbio.00047-21
- 4.Filkins LM, Graber JA, Olson DG, et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. DiRita VJ, ed. J Bacteriol. Published online July 15, 2015:2252-2264. doi:10.1128/jb.00059-15
- 5.Limoli DH, Whitfield GB, Kitao T, et al. Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. Harwood CS, ed. mBio. Published online May 3, 2017. doi:10.1128/mbio.00186-17
- 6.Trizna EY, Yarullina MN, Baidamshina DR, et al. Bidirectional alterations in antibiotics susceptibility in Staphylococcus aureus—Pseudomonas aeruginosa dual-species biofilm. Sci Rep. Published online September 9, 2020. doi:10.1038/s41598-020-71834-w
- 7.Rutherford ST, Bassler BL. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspectives in Medicine. Published online November 1, 2012:a012427-a012427. doi:10.1101/cshperspect.a012427
- 8.Uzi‐Gavrilov S, Tik Z, Sabti O, Meijler MM. Chemical Modification of a Bacterial Siderophore by a Competitor in Dual‐Species Biofilms. Angew Chem Int Ed. Published online June 13, 2023. doi:10.1002/anie.202300585
- 9.Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. Published online August 2016:828-837. doi:10.1038/nbt.3597
- 10.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J Bacteriol. Published online January 15, 2005:554-566. doi:10.1128/jb.187.2.554-566.2005
- 11.Jaiyesimi OA, McAvoy AC, Fogg DN, Garg N. Metabolomic profiling of Burkholderia cenocepacia in synthetic cystic fibrosis sputum medium reveals nutrient environment-specific production of virulence factors. Sci Rep. Published online November 1, 2021. doi:10.1038/s41598-021-00421-4
- 12.Jenul C, Keim K, Jens J, et al. Pyochelin biotransformation byStaphylococcus aureusshapes bacterial competition withPseudomonas aeruginosain polymicrobial infections. Published online April 18, 2022. doi:10.1101/2022.04.18.486787
- 13.Dührkop K, Fleischauer M, Ludwig M, et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. Published online March 18, 2019:299-302. doi:10.1038/s41592-019-0344-8
- 14.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. Published online September 21, 2015:12580-12585. doi:10.1073/pnas.1509788112
- 15.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. Published online September 12, 2007:211-221. doi:10.1007/s11306-007-0082-2
- 16.Pierson LS III, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. Published online March 30, 2010:1659-1670. doi:10.1007/s00253-010-2509-3
- 17.Vilaplana L, Marco MP. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: synthesis regulation, pathogenesis and analytical methods for their detection. Anal Bioanal Chem. Published online May 27, 2020:5897-5912. doi:10.1007/s00216-020-02696-4
- 18.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-Carboxylic Acid Promotes Bacterial Biofilm Development via Ferrous Iron Acquisition. J Bacteriol. Published online July 15, 2011:3606-3617. doi:10.1128/jb.00396-11
- 19.Wang Y, Newman DK. Redox Reactions of Phenazine Antibiotics with Ferric (Hydr)oxides and Molecular Oxygen. Environ Sci Technol. Published online February 20, 2008:2380-2386. doi:10.1021/es702290a
- 20.Pagmadulam B, Tserendulam D, Rentsenkhand T, et al. Isolation and characterization of antiprotozoal compound-producing Streptomyces species from Mongolian soils. Parasitology International. Published online February 2020:101961. doi:10.1016/j.parint.2019.101961
- 21.Xiong Z, Niu J, Liu H, Xu Z, Li J, Wu Q. Synthesis and bioactivities of Phenazine-1-carboxylic acid derivatives based on the modification of PCA carboxyl group. Bioorganic & Medicinal Chemistry Letters. Published online May 2017:2010-2013. doi:10.1016/j.bmcl.2017.03.011
- 22.Puopolo G, Masi M, Raio A, et al. Insights on the susceptibility of plant pathogenic fungi to phenazine-1-carboxylic acid and its chemical derivatives. Natural Product Research. Published online June 2013:956-966. doi:10.1080/14786419.2012.696257
- 23.Udumula V, Endres JL, Harper CN, et al. Simple synthesis of endophenazine G and other phenazines and their evaluation as anti-methicillin-resistant Staphylococcus aureus agents. European Journal of Medicinal Chemistry. Published online January 2017:710-721. doi:10.1016/j.ejmech.2016.09.079