Allergic Asthma: when your immune system is overreactive
Asthma is a prevalent chronic condition in children in developed countries, and it is known for its highly variable presentation and clinical course. Unfortunately, there is no well-established diagnostic gold standard that is easy to apply1. The numbers of misdiagnosis are high, both underdiagnosis, and overdiagnosis. Allergic asthma is the most common asthma phenotype in children. The immune system of patients is over-sensitive and reacts to common allergens such as pollen or mold, causing inflammation in the airways or making existing inflammation even worse, leading to hyper-reaction. Exposure to allergens can cause allergy-induced asthma attacks. Improving the diagnosis and management of pediatric allergic asthma is a crucial need. To address this, breath analysis aims to non-invasively evaluate altered metabolism and disease-associated processes. By identifying exhaled metabolic signatures that distinguish children with allergic asthma from healthy controls, clinicians can more accurately diagnose and manage pediatric asthma.
SESI/HRMS: breathing into the mass spectrometer
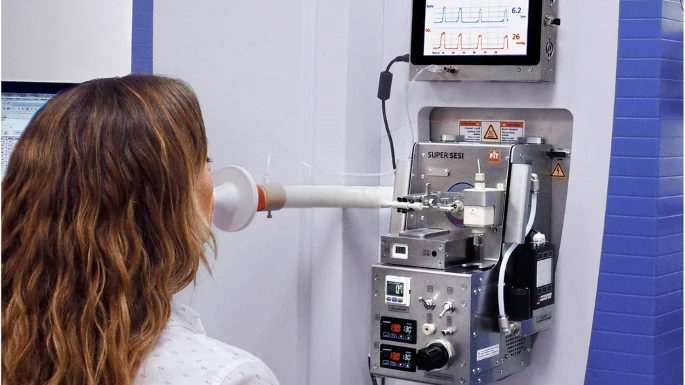
Secondary electrospray ionisation high-resolution mass spectrometry (SESI/HRMS) is a powerful method for the detection of volatile compounds without the need for sample pretreatment. SESI ionization works by ionizing volatile analytes through proton transfer reactions between the electrospray solution and the sample. This method is particularly useful for online breath analysis, allowing real-time measurements without sample preparation3. Previous studies have demonstrated the potential of SESI/HRMS for identifying relevant exhaled organic compounds and for revealing altered molecular pathways associated with various respiratory diseases3–5. Note that the method is limited by the lacking chromatographic separation, which hinders the distinction of isomeric compounds.
Detecting asthma from breath in children
In a cross-sectional study, researchers from the University Children’s Hospital Zurich, the University Hospital Zurich and the ETH Zurich collected exhaled breath samples from 48 allergic asthmatic children and 56 healthy control children6. All children were exhaling directly into the instrument (SuperSESI, FIT FossilionTech connected to a TripleTOF 5600+, AB Sciex). The researchers aimed to detect differentially expressed m/z features between the asthma patients and the control group. From a total of 2,315 m/z features detected in exhaled breath, 375 features exhibited significant differences between the two groups. Among these, 179 m/z features were upregulated, while 196 were downregulated in the allergic asthma group when compared to the control group.
Annotating relevant features with CSI:FIngerID
For the 100 most significantly discriminative m/z features per study group, the researchers employed MS2 spectra analysis, which were also directly recorded from the exhaled breath. They used SIRIUS+CSI:FingerID7,8 to assign putative molecular formulae and chemical structures. They further screened the putatively identified within their biological context and subgrouped into metabolic pathways or chemical families. Additionally, pathway enrichment analysis9 was performed to further aid in the identification of all significant features, including those without MS2 spectra or compound annotation. In total, 134 m/z features were successfully assigned to compounds. The certainty of identification ranges from level 1 (confirmed structure with measured reference standard) to level 5 (exact mass), with CSI:FingerID identifications being level 3 (tentative candidates)10
The pathophysiology of allergic asthma
Aim of this study was not only the identification of an asthmatic breath profile, but also to link the identified molecules to relevant pathways and compound classes that are associated with the pathophysiology of allergic asthma.
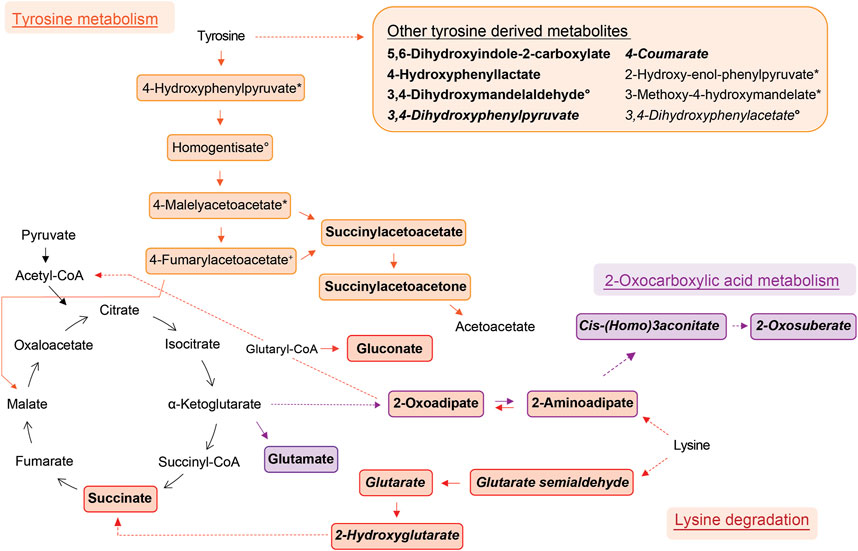
Elevated in allergic asthma:
- The researchers found the lysine metabolism to be the most significantly elevated pathway in asthmatic breath. Within this pathway, they unambiguously identified succinate and glutarate, which have been previously reported as associated with pediatric asthma in metabolomic studies using blood11, urine12, and breath13 samples.
- Also tyrosine metabolism is elevated. Previous metabolomic studies have reported increased levels of tyrosine in asthmatic children12,14,15, and high levels of tyrosine metabolism may be linked to inflammation and oxidative stress in asthma16
- Fatty acid metabolites constitute the largest elevated group, with 20 different fatty acid metabolites showing elevated levels. All lysine metabolites also belong to this chemical family. These fatty acid metabolites have been previously reported to be decreased in chronic obstructive pulmonary disease exacerbations3,17.
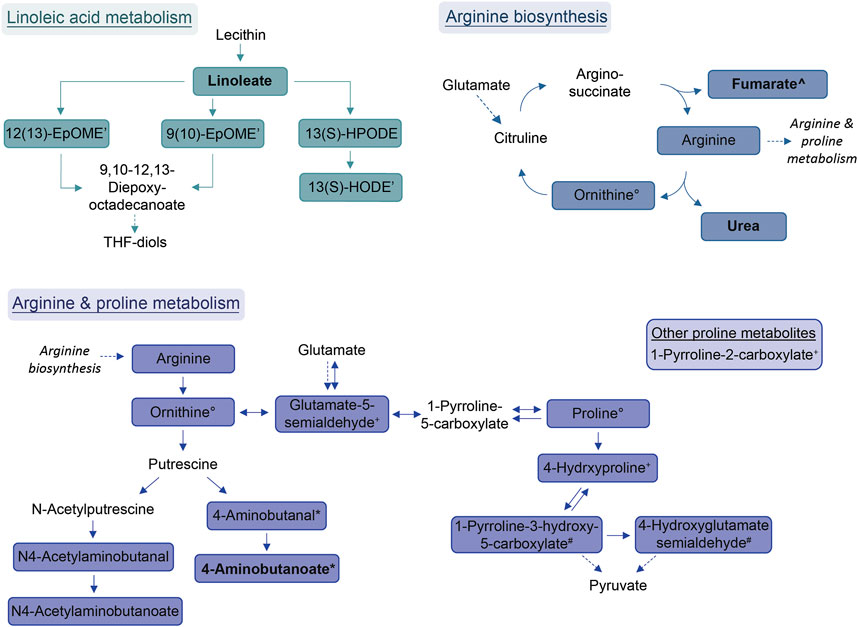
Diminished in allergic asthma:
- Arginine and proline metabolism, including arginine biosynthesis, are found to be diminished. Arginase is an important contributor to asthma pathophysiology18.
- Linoleic acid metabolism is another pathway that is diminished in allergic asthma. Linoleic acid has been reported to possess anti-inflammatory properties, but its effect on asthma remains controversial due to conflicting observations in clinical trials19. However, genetically predicted linoleic acid has been associated with a lower risk for asthma20.
- Additionally, amides, including palmitoylethanolamide, are another group of compounds showing diminished levels in allergic asthma. Palmitoylethanolamide, in particular, is well-studied for its anti-inflammatory effects21.
Do you have allergic asthma?
The researchers assess the ability of breath profiles to classify samples as either allergic asthmatic or healthy using support vector machines (SVM)22, a popular machine learning technique. The SVM algorithm was trained and tested using a 10 times repeated stratified 10-fold cross-validation approach. The resulting classifier has an area under the receiver operating characteristic (ROC) curve of 0.83, which is a reasonably high predictive accuracy in distinguishing between allergic asthmatic and healthy breath samples. The metabolic profiles derived from breath analysis could have potential diagnostic applications in the context of allergic asthma.
During the cross-validation process, certain compounds frequently emerged as significant predictors. This small group of compounds not only demonstrates potential relevance to the pathophysiology of allergic asthma but also holds promise for integration into diagnostic models. Specifically, the two dicarboxylic acids, succinate and glutarate, as well as lysine metabolites, were among the promising candidates and were unambiguously identified.
New path for improved asthma diagnostics
This is the first online breath analysis study to detect pediatric allergic asthma. The researchers identified a large number of discriminative features, many of which were putatively identified using the SIRIUS+CSI:FingerID. Additionally, they identified a small subset of compounds that exhibited strong differentiating power, suggesting their potential use in predictive modeling for asthma diagnosis. The study highlights the potential of breath analysis
- to provide valuable insights into the pathophysiology of allergic asthma in children,
- to uncover unique metabolic signatures associated with the disease, and
- to distinguishing children with allergic asthma from healthy controls.
This non-invasive approach shows promise for developing diagnostic tools that can aid in early detection and monitoring of asthma in children.
References
- 1.Gaillard EA, Kuehni CE, Turner S, et al. European Respiratory Society clinical practice guidelines for the diagnosis of asthma in children aged 5–16 years. Eur Respir J. Published online April 16, 2021:2004173. doi:10.1183/13993003.04173-2020
- 2.Singh KD, Tancev G, Decrue F, et al. Standardization procedures for real-time breath analysis by secondary electrospray ionization high-resolution mass spectrometry. Anal Bioanal Chem. Published online April 15, 2019:4883-4898. doi:10.1007/s00216-019-01764-8
- 3.Gaugg MT, Nussbaumer-Ochsner Y, Bregy L, et al. Real-Time Breath Analysis Reveals Specific Metabolic Signatures of COPD Exacerbations. Chest. Published online August 2019:269-276. doi:10.1016/j.chest.2018.12.023
- 4.Schwarz EI, Martinez-Lozano Sinues P, Bregy L, et al. Effects of CPAP therapy withdrawal on exhaled breath pattern in obstructive sleep apnoea. Thorax. Published online December 15, 2015:110-117. doi:10.1136/thoraxjnl-2015-207597
- 5.Weber R, Haas N, Baghdasaryan A, et al. Volatile organic compound breath signatures of children with cystic fibrosis by real-time SESI-HRMS. ERJ Open Res. Published online January 2020:00171-02019. doi:10.1183/23120541.00171-2019
- 6.Weber R, Streckenbach B, Welti L, et al. Online breath analysis with SESI/HRMS for metabolic signatures in children with allergic asthma. Front Mol Biosci. Published online March 31, 2023. doi:10.3389/fmolb.2023.1154536
- 7.Dührkop K, Fleischauer M, Ludwig M, et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. Published online March 18, 2019:299-302. doi:10.1038/s41592-019-0344-8
- 8.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. Published online September 21, 2015:12580-12585. doi:10.1073/pnas.1509788112
- 9.Pang Z, Chong J, Zhou G, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Research. Published online May 21, 2021:W388-W396. doi:10.1093/nar/gkab382
- 10.Schymanski EL, Jeon J, Gulde R, et al. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ Sci Technol. Published online February 18, 2014:2097-2098. doi:10.1021/es5002105
- 11.Chang C, Guo Z guo, He B, Yao W zhen. Metabolic alterations in the sera of Chinese patients with mild persistent asthma: a GC-MS-based metabolomics analysis. Acta Pharmacol Sin. Published online November 2015:1356-1366. doi:10.1038/aps.2015.102
- 12.Saude EJ, Skappak CD, Regush S, et al. Metabolomic profiling of asthma: Diagnostic utility of urine nuclear magnetic resonance spectroscopy. Journal of Allergy and Clinical Immunology. Published online March 2011:757-764.e6. doi:10.1016/j.jaci.2010.12.1077
- 13.Carraro S, Bozzetto S, Giordano G, et al. Wheezing preschool children with early-onset asthma reveal a specific metabolomic profile. Pediatr Allergy Immunol. Published online April 15, 2018:375-382. doi:10.1111/pai.12879
- 14.Papamichael MM, Katsardis C, Erbas B, Itsiopoulos C, Tsoukalas D. Urinary organic acids as biomarkers in the assessment of pulmonary function in children with asthma. Nutrition Research. Published online January 2019:31-40. doi:10.1016/j.nutres.2018.10.004
- 15.Tao J, Chen Y, Dai Q, et al. Urine metabolic profiles in paediatric asthma. Respirology. Published online February 14, 2019:572-581. doi:10.1111/resp.13479
- 16.Papamichael MM, Katsardis C, Sarandi E, et al. Application of Metabolomics in Pediatric Asthma: Prediction, Diagnosis and Personalized Treatment. Metabolites. Published online April 18, 2021:251. doi:10.3390/metabo11040251
- 17.Gaugg MT, Bruderer T, Nowak N, et al. Mass-Spectrometric Detection of Omega-Oxidation Products of Aliphatic Fatty Acids in Exhaled Breath. Anal Chem. Published online September 14, 2017:10329-10334. doi:10.1021/acs.analchem.7b02092
- 18.Maarsingh H, Dekkers BGJ, Zuidhof AB, et al. Increased arginase activity contributes to airway remodelling in chronic allergic asthma. European Respiratory Journal. Published online February 10, 2011:318-328. doi:10.1183/09031936.00057710
- 19.Wendell SG, Baffi C, Holguin F. Fatty acids, inflammation, and asthma. Journal of Allergy and Clinical Immunology. Published online May 2014:1255-1264. doi:10.1016/j.jaci.2013.12.1087
- 20.Zhao JV, Schooling CM. The role of linoleic acid in asthma and inflammatory markers: a Mendelian randomization study. The American Journal of Clinical Nutrition. Published online September 2019:685-690. doi:10.1093/ajcn/nqz130
- 21.Clayton P, Hill M, Bogoda N, Subah S, Venkatesh R. Palmitoylethanolamide: A Natural Compound for Health Management. IJMS. Published online May 18, 2021:5305. doi:10.3390/ijms22105305
- 22.Cortes C, Vapnik V. Support-vector networks. Mach Learn. Published online September 1995:273-297. doi:10.1007/bf00994018








