The Uncharted World of Metabolomics
Despite rapid advances in mass spectrometry hardware and co§mputational methods, confidently identifying the vast majority of metabolites detected in complex samples remains a challenging aspect of metabolomic research. On average, less than 10% of features are confidently annotated, leaving a vast, uncharted chemical space1. The core of this problem lies in the limited coverage of spectral libraries2 and the difficulty of predicting how unknown metabolites will fragment in a mass spectrometer.
Multiplexed Chemical Metabolomics: Chemical labeling to gain information
A research team led by Daniel Petras and Chambers C. Hughes at the University of Tübingen, in collaboration with the SIRIUS team, has developed a new workflow to address this challenge. Their new method, Multiplexed Chemical Metabolomics (MCheM), integrates online chemical labeling reactions directly into the liquid chromatography-tandem mass spectrometry (LC-MS/MS) workflow3.
The MCheM workflow uses an array of post-column derivatisation reactions to generate additional structural information. Samples are separated by liquid chromatography (LC), and then, just before they enter the mass spectrometer, specific chemical agents are continuously infused to react with particular functional groups on the metabolites and generate derivatised versions of the molecules.
The current MCheM workflow employs three key reactions:
- L-cysteine targets electrophiles.
- 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC) targets amino and phenol groups.
- Hydroxylamine hydrochloride targets aldehydes and ketones.
The advantage over traditional batch derivatisation methods4, is that the link between the precursor molecule and its derivatisation product is maintained throughout the analysis. Due to its simple hardware setup, MCheM is easily implemented on most commercial LC-MS/MS platforms, making it broadly applicable.
Restricting the molecular structure search space in SIRIUS
The true power of MCheM emerges when this rich, reactivity-based information is integrated with advanced computational tools. The “Online Reactivity” analysis module in mzmine leverages the co-elution of precursors and products to establish correlation-based connections, using ion identity networking5,6. This creates a hybrid dataset that integrates MS, MS/MS, and reactivity-based information.
This integrated data, which includes a list of predicted functional groups or substructures in the form of SMILES Arbitrary Target Specification (SMARTS), can be directly imported to SIRIUS to improve annotation confidence. By incorporating MCheM-derived functional group information, SIRIUS can constrain the molecular structure search space, leading to far more accurate and confident metabolite annotations. SIRIUS structure identification results can be filtered to show only results that fit into MCheM-derived constraints.
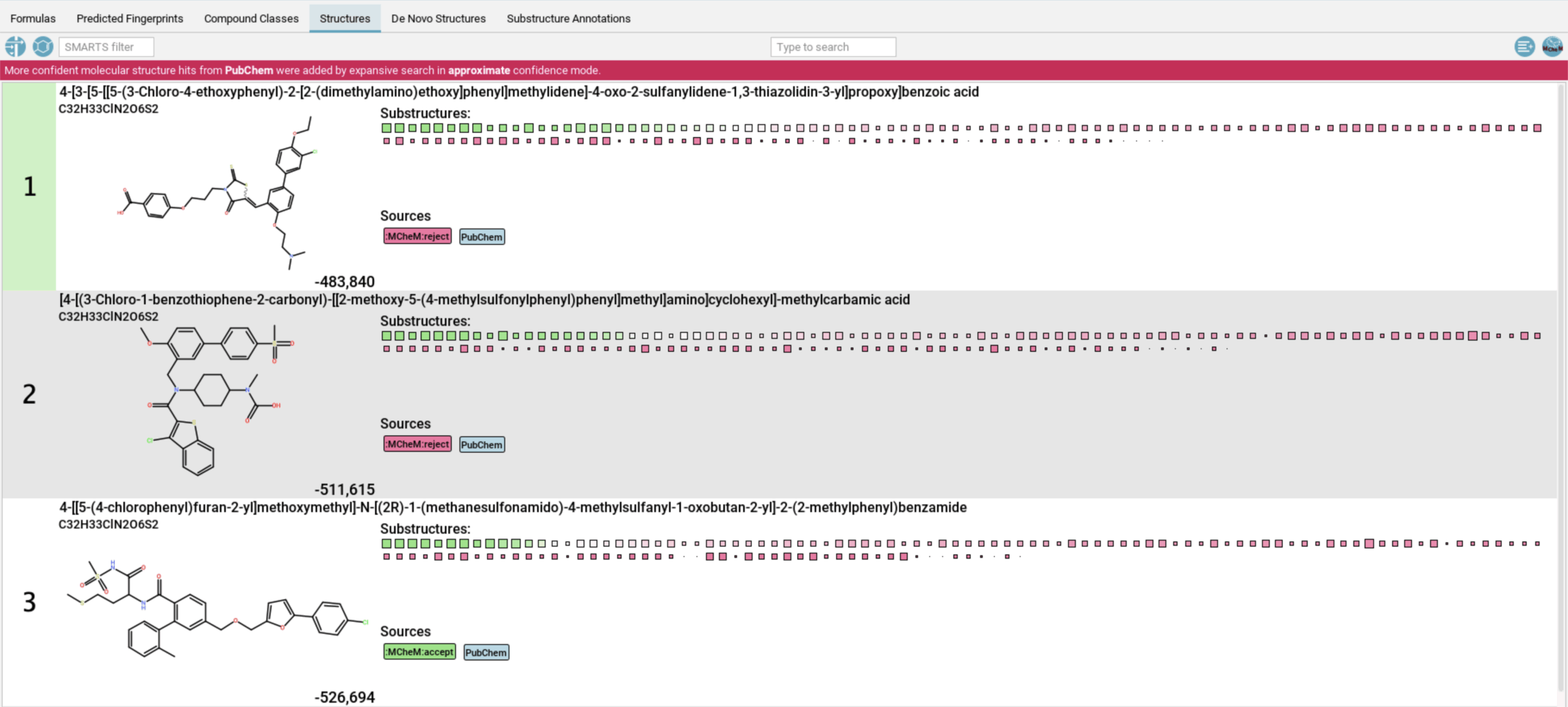
When analyzing an experimental library of about 10,000 compounds, MCheM-enhanced data improved the SIRIUS results for 32% of the molecules, with 15% improving to a top-1 match. For a set of authentic natural product standards, annotation improvements were even higher (49%).
Accelerating discovery of novel compounds
The impact of MCheM extends beyond just better numbers; it directly facilitates the discovery of novel compounds. In a genome-guided natural product discovery case study, the team used the workflow to explore uncharacterized bacterial extracts from Streptomyces libani subsp. rufus DSM 41230, a microbe known for producing natural products. The S. libani genome contains a biosynthetic gene cluster (BGC) similar to the oxazolomycin B gene cluster, a family of natural products that includes a reactive β-lactone moiety7. The MCheM workflow was specifically employed to detect this moiety using the L-cysteine reaction.
Using MCheM-based reranking of CSI:FingerID search results, oxazolomycin D was identified as the top annotation for m/z 700.3804, which had not been readily annotated as an oxazolomycin derivative using regular MS/MS data alone. This demonstrates MCheM’s effectiveness in improving the identification of natural products. Further analysis led to the identification and structure elucidation of 7-glycosyl oxazolomycin D, a previously undescribed and unlisted member of the oxazolomycin family. The novel structure was confirmed using a suite of orthogonal NMR experiments, highlighting the power of MCheM to guide the discovery of truly novel molecules.
Broadening the Horizons of Metabolomics
MCheM represents a significant leap forward in metabolomics, offering a powerful new toolbox for the structure elucidation of unknown metabolites at scale. By integrating sophisticated chemical derivatisation with the analytical powers of SIRIUS, the potential for discovery grows even greater as more functional groups are targeted. This approach offers a scalable and accessible toolkit for expanding our understanding of the vast and uncharted chemical space of metabolites.
Giovanni Andrea Vitale, Shu-Ning Xia, Kai Dührkop, Mohammad Reza Zare Shahneh, Heike Brötz-Oesterhelt, Yvonne Mast, Corinna Brungs, Sebastian Böcker, Robin Schmid, Mingxun Wang, Chambers C. Hughes & Daniel Petras.
Enhancing tandem mass spectrometry-based metabolite annotation with online chemical labeling.
Nat Commun. 2025 Jul 26;16(1):6911. doi: 10.1038/s41467-025-61240-z
References
- Bittremieux W, Wang M, Dorrestein PC. The critical role that spectral libraries play in capturing the metabolomics community knowledge. Metabolomics. 2022 Nov 19;18(12):94. doi: 10.1007/s11306-022-01947-y. ↩︎
- Theodoridis G, Gika H, Raftery D, Goodacre R, Plumb RS, Wilson ID. Ensuring Fact-Based Metabolite Identification in Liquid Chromatography-Mass Spectrometry-Based Metabolomics. Anal Chem. 2023 Feb 28;95(8):3909-3916. doi: 10.1021/acs.analchem.2c05192. ↩︎
- Vitale GA, Xia SN, Dührkop K, Zare Shahneh MR, Brötz-Oesterhelt H, Mast Y, Brungs C, Böcker S, Schmid R, Wang M, Hughes CC, Petras D. Enhancing tandem mass spectrometry-based metabolite annotation with online chemical labeling. Nat Commun. 2025 Jul 26;16(1):6911. doi: 10.1038/s41467-025-61240-z. ↩︎
- Kaur A, Lin W, Dovhalyuk V, Driutti L, Di Martino ML, Vujasinovic M, Löhr JM, Sellin ME, Globisch D. Chemoselective bicyclobutane-based mass spectrometric detection of biological thiols uncovers human and bacterial metabolites. Chem Sci. 2023 Apr 6;14(20):5291-5301. doi: 10.1039/d3sc00224a. ↩︎
- Schmid R, Heuckeroth S, Korf A, Smirnov A, Myers O, Dyrlund TS, Bushuiev R, Murray KJ, Hoffmann N, Lu M, Sarvepalli A, Zhang Z, Fleischauer M, Dührkop K, Wesner M, Hoogstra SJ, Rudt E, Mokshyna O, Brungs C, Ponomarov K, Mutabdžija L, Damiani T, Pudney CJ, Earll M, Helmer PO, Fallon TR, Schulze T, Rivas-Ubach A, Bilbao A, Richter H, Nothias LF, Wang M, Orešič M, Weng JK, Böcker S, Jeibmann A, Hayen H, Karst U, Dorrestein PC, Petras D, Du X, Pluskal T. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat Biotechnol. 2023 Apr;41(4):447-449. doi: 10.1038/s41587-023-01690-2. ↩︎
- Schmid R, Petras D, Nothias LF, Wang M, Aron AT, Jagels A, Tsugawa H, Rainer J, Garcia-Aloy M, Dührkop K, Korf A, Pluskal T, Kameník Z, Jarmusch AK, Caraballo-Rodríguez AM, Weldon KC, Nothias-Esposito M, Aksenov AA, Bauermeister A, Albarracin Orio A, Grundmann CO, Vargas F, Koester I, Gauglitz JM, Gentry EC, Hövelmann Y, Kalinina SA, Pendergraft MA, Panitchpakdi M, Tehan R, Le Gouellec A, Aleti G, Mannochio Russo H, Arndt B, Hübner F, Hayen H, Zhi H, Raffatellu M, Prather KA, Aluwihare LI, Böcker S, McPhail KL, Humpf HU, Karst U, Dorrestein PC. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat Commun. 2021 Jun 22;12(1):3832. doi: 10.1038/s41467-021-23953-9. ↩︎
- Zhao C, Ju J, Christenson SD, Smith WC, Song D, Zhou X, Shen B, Deng Z. Utilization of the methoxymalonyl-acyl carrier protein biosynthesis locus for cloning the oxazolomycin biosynthetic gene cluster from Streptomyces albus JA3453. J Bacteriol. 2006 Jun;188(11):4142-7. doi: 10.1128/JB.00173-06. ↩︎