About us

Our Vision
We want to establish the identification of unknown small molecules as a standard method in various industries: for drug discovery, for the detection of contaminants in various products and foods, for water analysis, or as a doping test. Even today, the majority of small molecules remain unexplored, yet unlocking this knowledge is fundamental to various realms of research and industry. Our cutting-edge software solutions serve as the foundation for shaping the future of small molecule structure elucidation.

Our History
Bright Giant GmbH was established in 2019 as a spin-off from the Friedrich Schiller University Jena, Germany. The algorithms underlying our software have been developed in years of academic research. We have transferred our software to the business sector optimising it for maximum usability and providing the requisite support to seamlessly integrate research-oriented software into large enterprises. At the same time, we stay strongly connected to academic research and the academic community benefits from our continuous improvements of SIRIUS.

Our Customers
Justified by the interest of numerous companies in the developed software solutions, our company has transferred SIRIUS to the business sector. We have customers from a wide range of sectors. These include leading international pharmaceutical and food companies, emerging biotech companies, but also service providers of analytical measurements.
Our Software and Services
SIRIUS open source project
Together with the University of Jena, we are developing our open source project SIRIUS, a java-based software framework for the de novo identification of metabolites and other small molecules of biological interest from LC-MS/MS data.
Licences
Bright Giant GmbH is the only license provider for SIRIUS web services and related SaaS solutions for non-academic users. The University of Jena offers similar SIRIUS web services free of charge to the academic community. Please review the terms of service of the academic version for details.
Custom services
We provide custom hosting of our software solutions, custom tailored workflows and reporting, custom structure database generation and custom training of machine learning models to include customer’s own reference data. Get your personalized structure elucidation workflow.
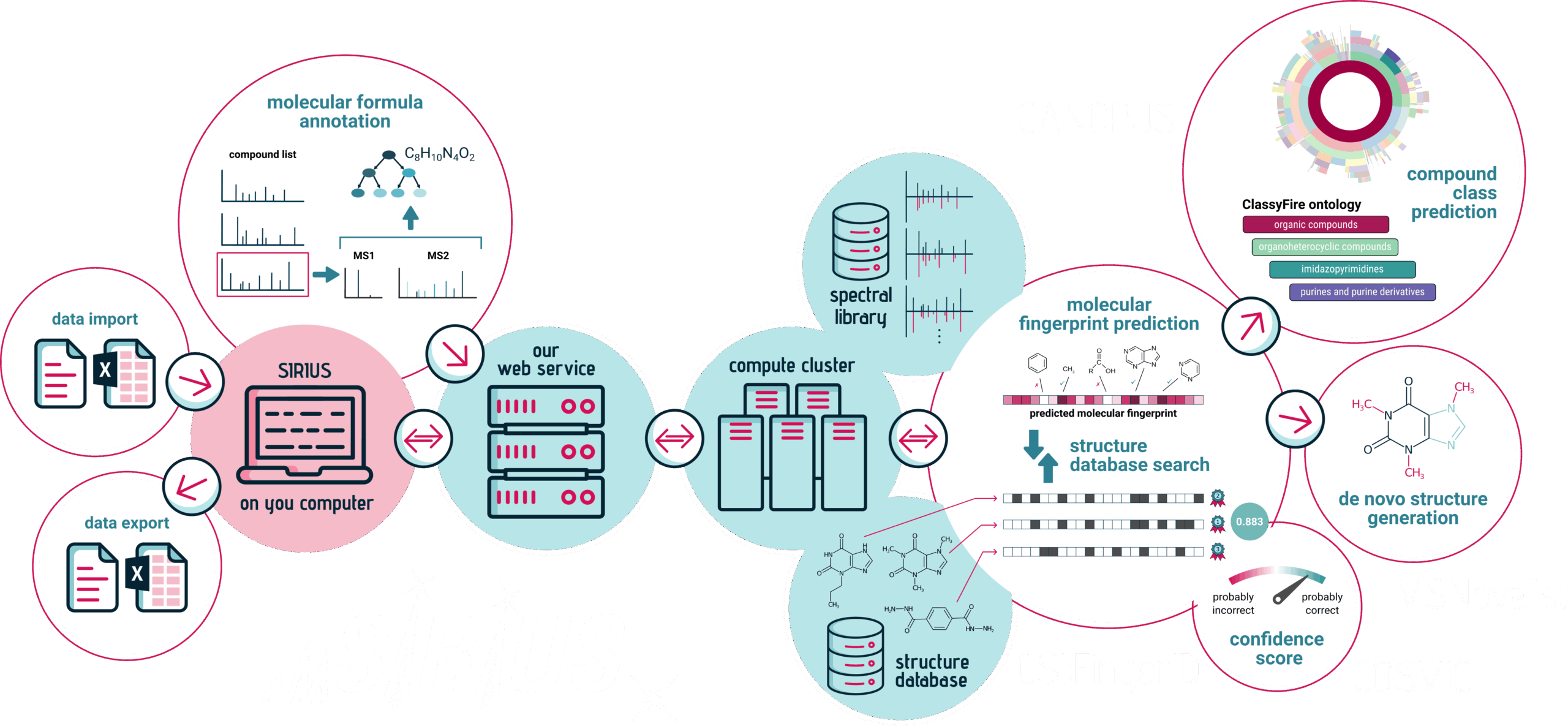
SIRIUS integrates a collection of algorithmic tools via the SIRIUS web services. In particular, both the graphical user interface and the command line version of SIRIUS seamlessly integrate the following tools.
CSI:FingerID
Molecular structure annotation
CSI:FingerID annotates unknown compounds from your fragmentation spectra by utilizing the fragmentation patterns of the molecules to search in databases of millions of molecular structures. Results are retrieved from the CSI:FingerID web service and can be displayed in the SIRIUS graphical user interface.
COSMIC
Identification confidence
The COSMIC confidence score assigns a confidence value to the CSI:FingerID structure annotations, allowing you to determine the likelihood of correct annotation. This is similar in spirit to what is done in spectral library search and enables automated high-throughput analysis of your mass spectrometry data.
CANOPUS
Compound class prediction
CANOPUS predicts over 2500 compound classes from your fragmentation spectra without relying on ANY database (not even structure databases).
MSNovelist
De novo structure generation
MSNovelist overcomes the limitations of database searches for unknown compounds by generating multiple novel candidate structures from a predicted molecular fingerprint and ranking them with CSI:FingerID.
Beyond the core algorithmic features, SIRIUS provides a comprehensive set of additional features streamlining the analysis workflow from feature detection to detailed result validation.
Beyond the core algorithmic features, SIRIUS provides a comprehensive set of additional features streamlining the analysis workflow from feature detection to detailed result validation.
Our Team

Dr. Marcus Ludwig
Marcus is the chief executive officer and a co-founder of Bright Giant. He earned his doctorate in computational mass spectrometry, working on algorithms for molecular formula and structure annotation. He has a long-standing experience in Bayesian optimization, solving small molecule annotation on whole-dataset level, and aims to advance such holistic approaches for small molecule annotation. As a part of Bright Giant, he focuses on business relations and funding opportunities.

Dr. Franziska Hufsky
Franziska joined Bright Giant in 2023 as the Scientific Outreach Coordinator. She received her PhD in computational mass spectrometry for small molecules and developed efficient algorithms to calculate and compare fragmentation trees from electron ionization spectra. She has many years of experience in outreach and equal opportunity, and has coordinated a research network for several years. On a personal note, she enjoys the outdoors biking or hiking, and loves books, coffee, and octopuses.

Dr. Markus Fleischauer
Markus is the chief technology officer and a co-founder of Bright Giant. During his PhD, he developed algorithms and software solutions for computational phylogenetics. He has a long-standing experience as a lead developer, coordinating the software development of SIRIUS and developing essential parts of the software and the web service infrastructure. Now he directs the development of the software solutions and the IT-Infrastructure at Bright Giant.

Dr. Kai Dührkop
Kai is co-founder of Bright Giant. He earned his doctorate in computational mass spectrometry, being the lead developer of CSI:FingerID. He has many years of experience in algorithm development and machine learning. At Bright Giant GmbH he directs the scientific development of innovative solutions in the field of algorithmic mass spectrometry of small molecules, ensuring that SIRIUS remains methodologically state-of-the-art.

Prof. Dr. Sebastian Böcker
Sebastian is co-founder of Bright Giant. He has over two decades of experience working on computational methods for the analysis of mass spectrometry data. Today, his lab at the Friedrich Schiller University Jena is one of the world-leading groups in algorithmic mass spectrometry for small molecules. The development of SIRIUS started in his lab. Since then, he and his team work consistently to ensure that the methods are not only best in class, but also as easy as possible for users to access. He brings his long-standing experience as a scientific advisor to Bright Giant.

Andrii Kovalov
Andrii joined Bright Giant in 2023 as a Senior Software Developer, bringing a decade of software engineering experience, and a passion for clean code, clean architecture and clean interfaces. He previously worked in the space sector, writing software for modelling satellites, and applying formal methods. In his spare time he loves rock climbing, playing guitar, and developing computer games.

Dr. Martin Hoffmann
Martin is the chief customer officer and a co-founder of Bright Giant. He did his PhD in computational mass spectrometry, focusing on confidence estimation of molecular structure predictions. As a part of Bright Giant GmbH, he manages the customer relations.

Dr. Martin Engler-Lukajewski
Martin joined Bright Giant in 2023 as a Senior Scientist. He received his PhD in computational mass spectrometry and algorithm engineering, developing a software framework to quantitatively analyze synthetic polymer sequences. Martin has a long-standing experience in software engineering, high-performance computing and machine learning and loves to write innovative and clean software to tackle complex problems. On a personal note, he is a Pratchett, Moers and Star Trek enthusiast.
Recognition
Awards
The algorithms underlying SIRIUS have been developed for many years in an academic context at the chair of Prof. Sebastian Böcker.
Prof. Sebastian Böcker, Dr. Kai Dührkop, Dr. Markus Fleischauer, Dr. Marcus Ludwig and Martin Hoffmann were awarded the Thuringian Research Prize 2022 for applied research.
SIRIUS was awarded “method to watch” by Nature Methods in 2020.
SIRIUS+CSI:FingerID has proven to be the best computational method to identify molecules from tandem mass spectrometry data by winning the CASMI contest for structure identification in 2016 and 2017 in category 2 (automatic structure identification).

Testimonials
Press Releases
mzio GmbH and Bright Giant GmbH Announce Seamless Integration Between mzmine and SIRIUS
mzio GmbH and Bright Giant GmbH announce a new integration between mzmine and SIRIUS. This collaboration enables researchers to launch SIRIUS analysis directly from mzmine feature tables, automating result handling and eliminating the need for manual data transfer to streamline small molecule discovery workflows.
References
Martin A. Hoffmann, Louis-Félix Nothias, Marcus Ludwig, Markus Fleischauer, Emily C. Gentry, Michael Witting, Pieter C. Dorrestein, Kai Dührkop and Sebastian Böcker. High-confidence structural annotation of metabolites absent from spectral libraries. Nature Biotechnology 40, 411–421, 2022.
Kai Dührkop, Louis-Félix Nothias, Markus Fleischauer, Raphael Reher, Marcus Ludwig, Martin A. Hoffmann, Daniel Petras, William H. Gerwick, Juho Rousu, Pieter C. Dorrestein and Sebastian Böcker. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nature Biotechnology, 39, 462–471, 2021.
Marcus Ludwig, Louis-Félix Nothias, Kai Dührkop, Irina Koester, Markus Fleischauer, Martin A. Hoffmann, Daniel Petras, Fernando Vargas, Mustafa Morsy, Lihini Aluwihare, Pieter C. Dorrestein, Sebastian Böcker. Database-independent molecular formula annotation using Gibbs sampling through ZODIAC. Nature Machine Intelligence 2, 629–641, 2020.
Kai Dührkop, Markus Fleischauer, Marcus Ludwig, Alexander A. Aksenov, Alexey V. Melnik, Marvin Meusel, Pieter C. Dorrestein, Juho Rousu, and Sebastian Böcker, SIRIUS 4: Turning tandem mass spectra into metabolite structure information. Nature Methods 16, 299–302, 2019.
Kai Dührkop and Sebastian Böcker. Fragmentation trees reloaded. Journal of Cheminformatics 8, 5, 2016.
Kai Dührkop, Huibin Shen, Marvin Meusel, Juho Rousu, and Sebastian Böcker. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proceedings of the National Academy of Sciences USA 112(41), 12580-12585, 2015.
Sebastian Böcker, Matthias C. Letzel, Zsuzsanna Lipták and Anton Pervukhin. SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics 25(2), 218-224, 2009.

