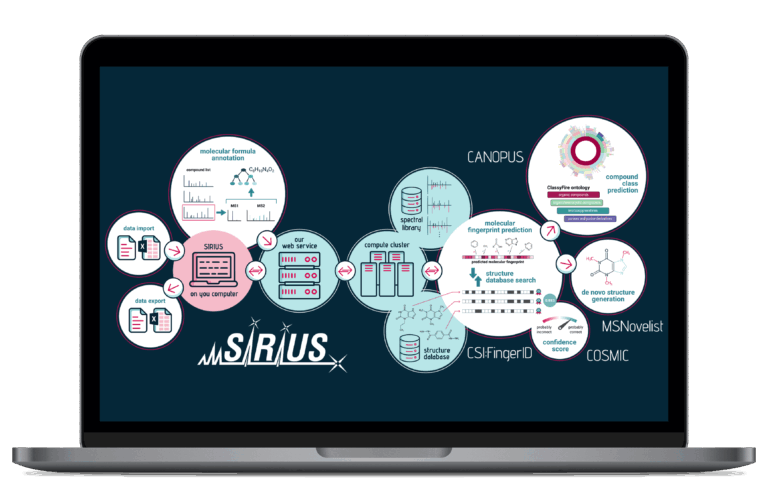
SIRIUS is the comprehensive software solution for the high-throughput identification of small molecules from fragmentation mass spectrometry data. It is engineered to address the inherent limitations of library-dependent identification methods. SIRIUS provides a comprehensive set of features spanning every step from feature detection to detailed result validation. It is designed to not only accurately characterize known compounds but also to confidently identify “unknown unknowns” in complex biological samples.
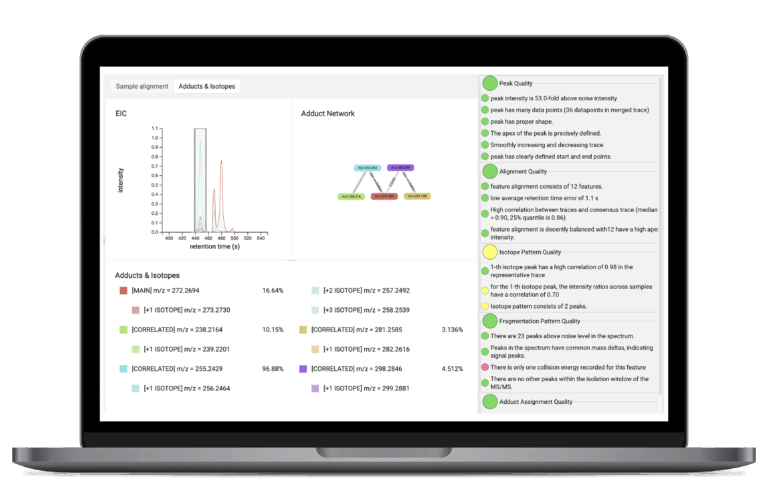
LC/MS Preprocessing
SIRIUS offers integrated LC/MS preprocessing tools that automate the essential steps of preparing your complex LC/MS data for high-throughput analysis. It can automatically extract and align chromatographic features across multiple samples, including robust feature detection and adduct detection. This process minimizes effort while providing associated quality metrics that enable users to effectively prioritize and filter results for downstream analysis.
Molecular Formula Annotation
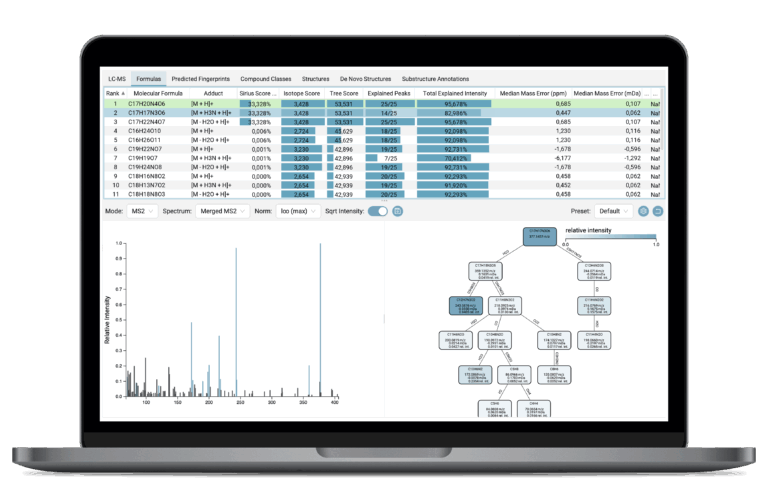
SIRIUS delivers accurate molecular formula annotation using a dual approach—analyzing both high-resolution isotope patterns and complex fragmentation patterns—to propose and score the most accurate elemental composition without needing a spectral library. It offers powerful search strategies for distinct use-cases, from generic applications to the discovery of “unknown unknowns”.
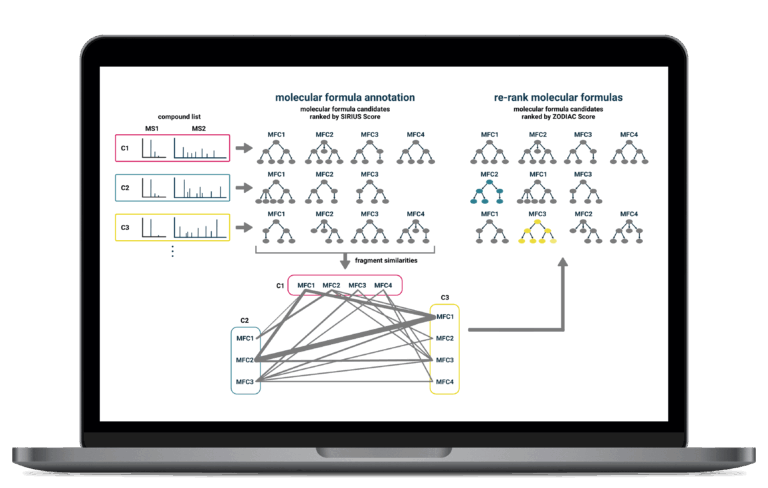
Network-Enhanced Molecular Formula Annotation
for Complex Datasets
For complex biological datasets, SIRIUS improves the accuracy of molecular formula annotations by analyzing relationships across the entire dataset. It uses a sophisticated network-based algorithm that leverages the fact that related compounds often share similarities in their biosynthetic pathways and fragmentation patterns.
Molecular Fingerprint Prediction
SIRIUS predicts a compound’s unique, machine-readable molecular fingerprint (a set of structural properties) from the fragmentation pattern. This is the crucial, intermediary feature that powers highly accurate structure and class prediction.
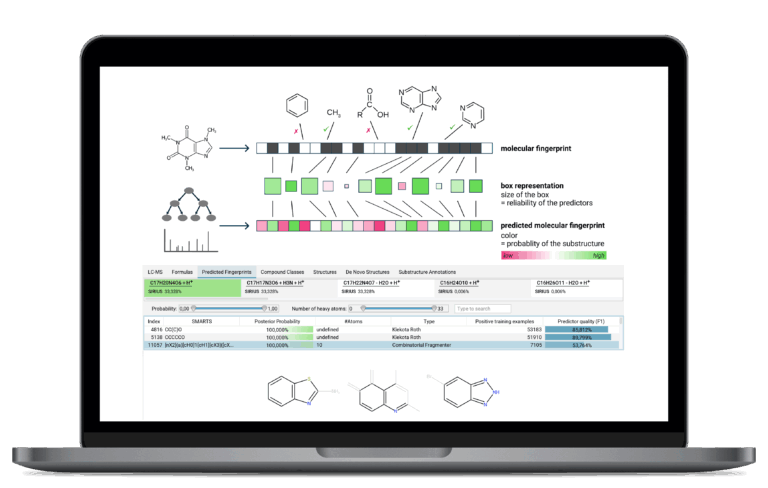
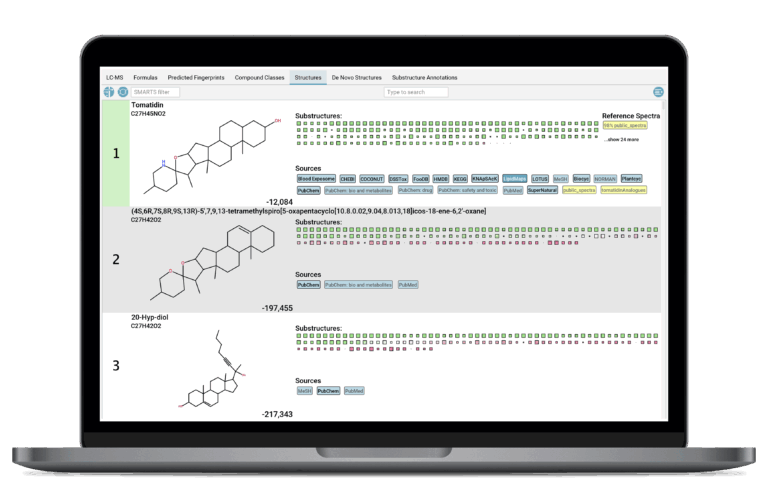
Molecular Structure Annotation
SIRIUS uses the molecular fingerprint to efficiently search massive molecular structure databases, such as PubChem, to identify the most likely structure candidate for your unknown compound. SIRIUS ships with a wide range of built-in databases. Additionally, users can enhance the search capabilities by adding custom databases with their own structures.
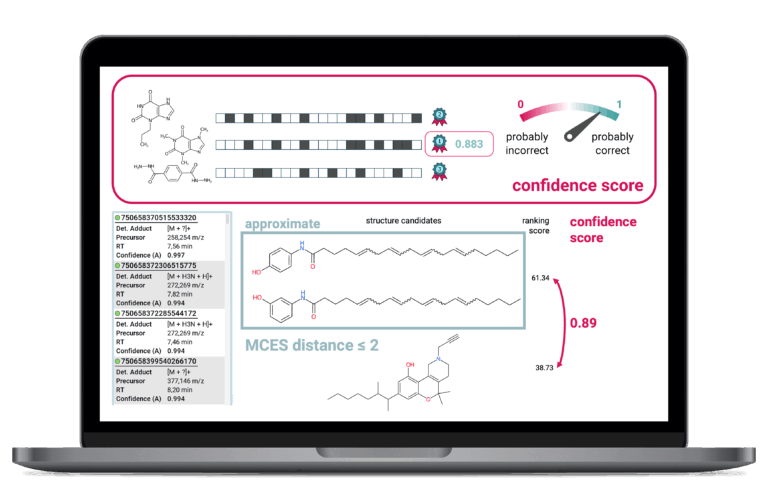
Confidence in Annotation Results
SIRIUS provides a confidence score for every structure prediction result that assigns a measure of reliability to your compound identification. Assessing the trustworthiness of results is essential for prioritizing hits in large-scale analysis. SIRIUS offers two distinct modes, allowing users to evaluate either the probability of the hit being the exact structure or the probability of it being highly similar to the true structure.
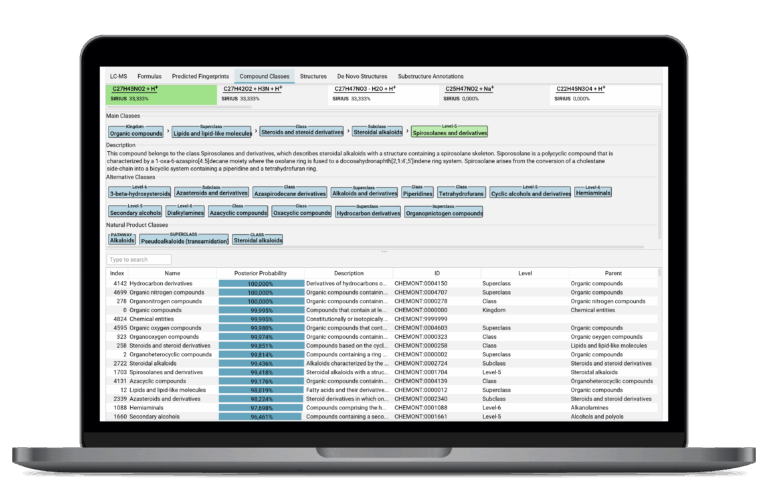
Chemical Classification
SIRIUS predicts the complete chemical class hierarchy (e.g., lipid, alkaloid, organoheterocyclic) of an unknown compound by assigning ClassyFire compound classes directly from the molecule’s predicted molecular fingerprint, requiring neither spectral nor structural reference data. This feature immediately provides contextual understanding even for novel or unannotated compounds.
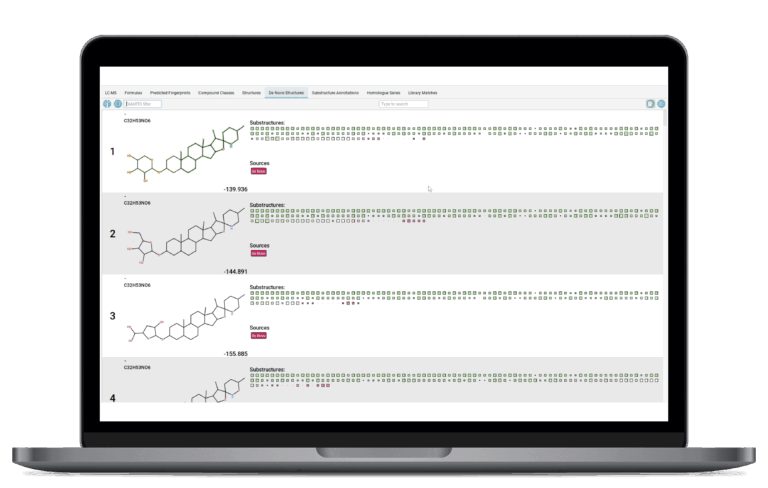
De Novo Structure Generation
SIRIUS empowers you to discover truly novel molecules by addressing the limitations of searching finite structure databases. Structures are generated from scratch based on the molecular formula and the predicted molecular fingerprint. This feature represents the next building block in structure elucidation, empowering you to explore the uncharacterized chemical space.
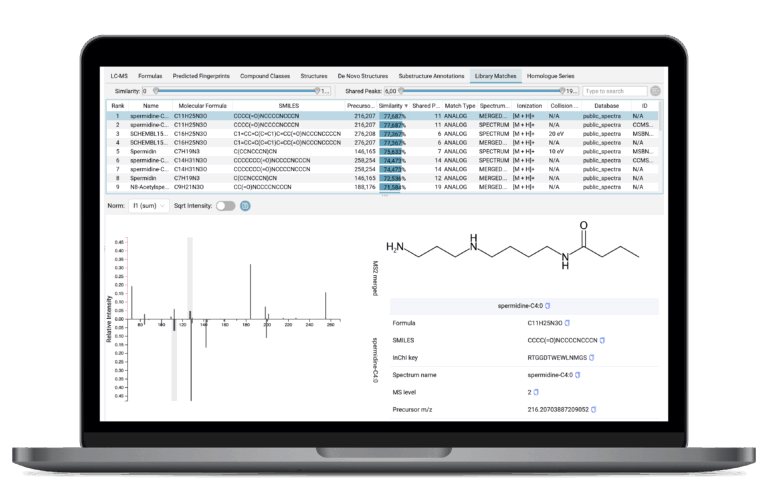
Spectral Library (Analog) Search
SIRIUS seamlessly combines powerful structure annotation tools with your in-house spectral libraries. It enables rapid matching and analog search that goes beyond identical matches to help you identify related compounds with similar core structures. These spectral library hits are auxiliary annotations alongside the main structural results, and can be leveraged by the user for result validation or by other SIRIUS tools—for instance, to strengthen molecular formula annotations by acting as anchors in the network-based analysis.
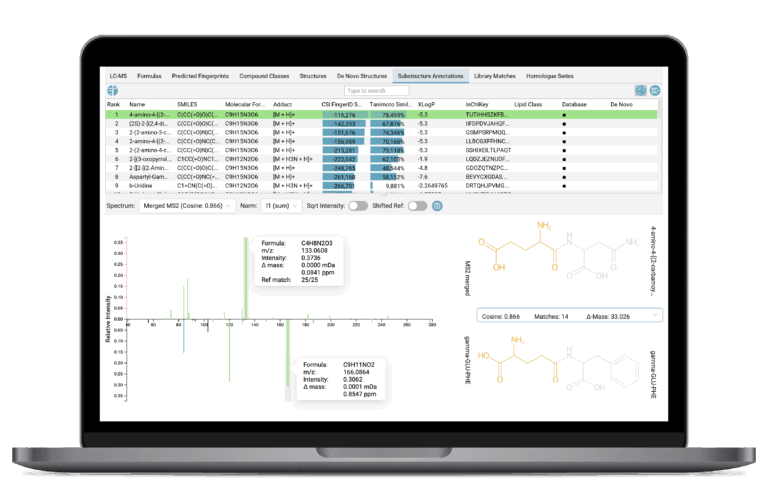
Substructure Validation
SIRIUS provides powerful visualization tools to facilitate the manual and objective assessment of structural assignments. Substructure highlighting allows you to see which predicted molecular features (substructures) support or contradict a candidate structure. Substructure annotations are based on the combinatorial fragmentation of the candidate structure and link specific fragment or neutral loss peaks in the measured spectrum to the corresponding proposed substructures of the candidate molecule. This is particularly helpful to validate structure hits for unknown compounds by highlighting shared substructure explanations observed in spectral library analogs.
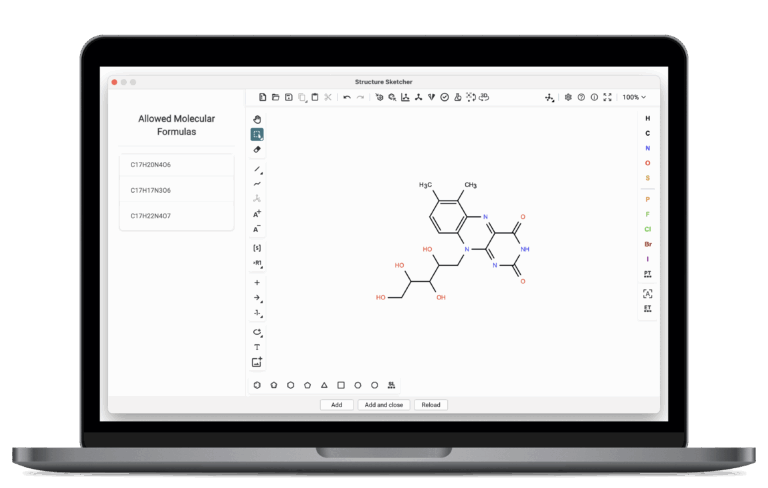
Structure Sketcher
SIRIUS provides an integrated chemistry workbench to quickly edit chemical structures directly within the software. You can manually modify existing candidate structures or submit new ones, and re-score them against predicted fingerprints to improve annotations with your chemical knowledge.
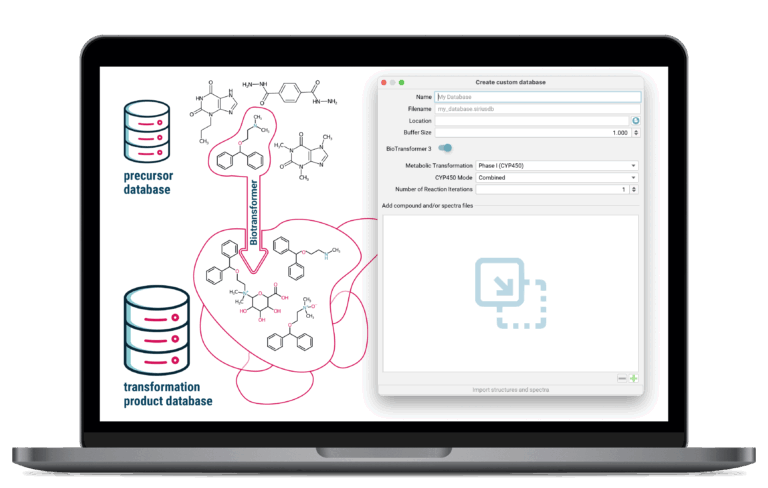
Biotransformer 3.0 Integration
SIRIUS seamlessly incorporates biotransformation predictions into your analysis. Generate custom, in silico databases of expected metabolic or degradation products on the fly, dramatically simplifying the identification of drug metabolites or environmental breakdown products.
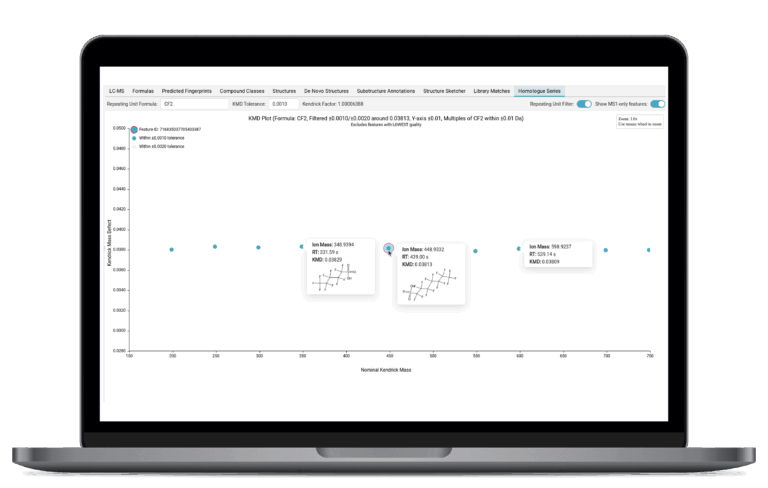
Kendrick Mass Defect Plots for Homologous Series
SIRIUS generates Kendrick Mass Defect plots—a powerful visualization tool that groups related molecules into homologous series. This simplifies the analysis of compounds that share a common core structure but differ by specific, repeating chemical units, making it easy to spot and confirm common structural patterns across your samples.
Powerful Suite of Interfaces
SIRIUS provides a powerful suite of interfaces: an intuitive Graphical User Interface (GUI) for visual analysis, a Command Line Interface (CLI) to automate large analysis, and an Application Programming Interface (API) to integrate SIRIUS into your custom pipelines. These interfaces share a persistence layer, allowing results from high-throughput computation (e.g., via CLI) to be manually inspected (via GUI).
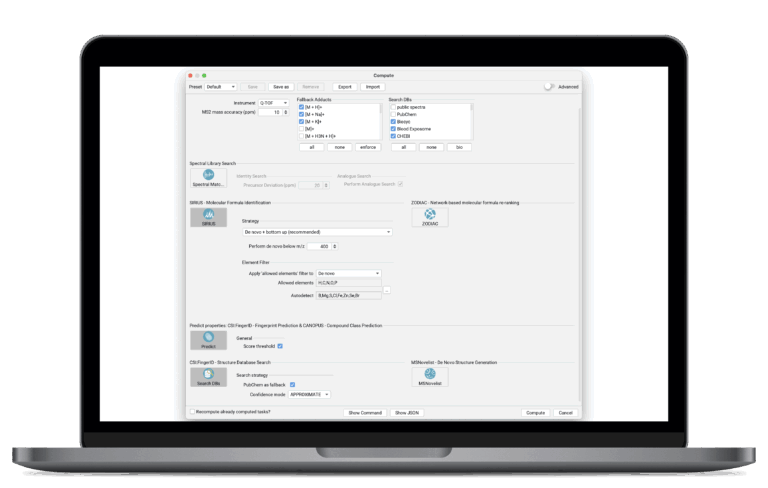
Fast, Reproducible Results with Streamlined Workflows
SIRIUS integrates all identification steps, from molecular formula annotation to structure and class prediction, into a coherent workflow. You can utilize expertly curated, one-click computation presets that deploy optimized, validated workflows of SIRIUS features for common analysis scenarios. Features auto-enable/disable based on workflow principles, with presets for saving and reloading setups.
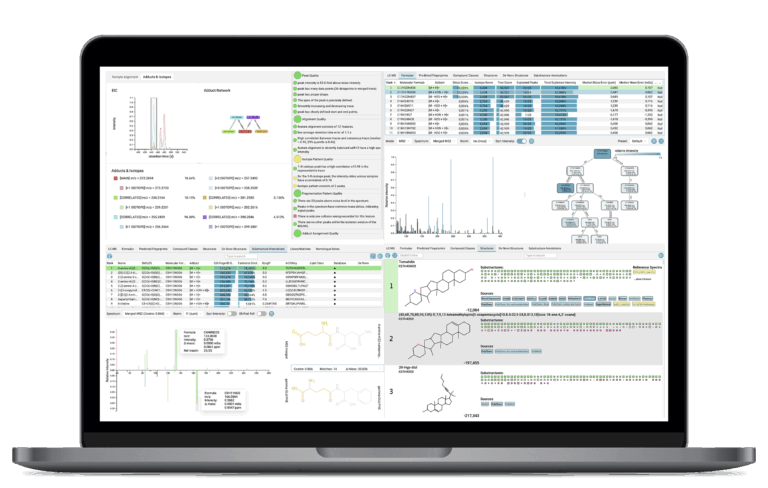
Visualizing Results
The SIRIUS graphical user interface offers several dedicated views to interpret results. Navigate systematic, organized views for every step of the process—formulas, compound classes, structures, and library matches—allowing for rapid, confident interpretation and reporting.
High-Throughput Processing of Full LC-MS Datasets
SIRIUS is designed for the high-throughput processing of full LC-MS datasets. Significant algorithm engineering and the implementation of a powerful job-scheduling system enable parallelization across multiple cores, reducing the time required to analyze a full run from hours to minutes.
SIRIUS is proven to be the best computational method for identifying molecules from fragmentation mass spectrometry data.
Move beyond the constraints of spectral libraries and download SIRIUS now to advance your high-throughput small molecule identification.

