Getting a structure database hit in SIRIUS is often just the beginning, not the final step, of annotation. While computational methods are powerful for generating initial hypotheses, the subsequent steps for structure elucidation involve crucial experimental validation and refinement. This is where your chemical knowledge becomes indispensable, allowing you to evaluate computational hypotheses and guide further investigation.
SIRIUS provides multiple layers for validating structure database search hits, for example compound class predictions, analogous spectral hits and substructure annotations. By combining this information with your chemical intuition, you can manually modify a structure candidate to significantly improve your annotation.
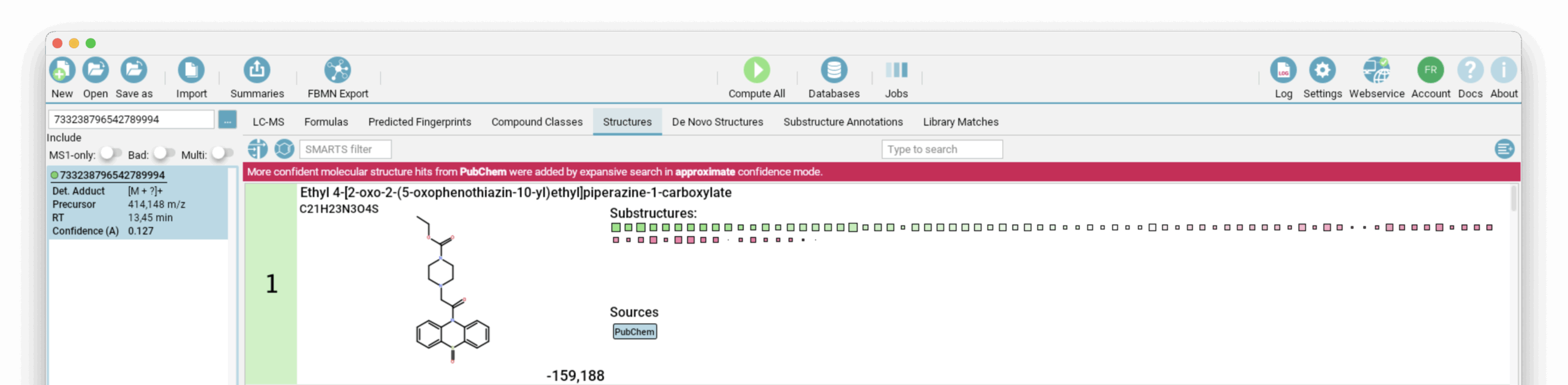
Let’s dive into an example. We have a feature at m/z 414.148, which we’ve searched against SIRIUS’s bio structure databases and a spectral library (for identity and analog hits). There wasn’t an identity hit in the spectral library, but we found several analog matches. Since no hits were found in the bio databases, we used PubChem as a fallback. The best hit from PubChem was Ethyl 4-[2-oxo-2-(5-oxophenothiazin-10-yl)ethyl]piperazine-1-carboxylate.
Identifying Potential Inaccuracies with SIRIUS’ Validation Tools
To better understand our feature and gain deeper insights into its molecular structure, we use different validation levels in SIRIUS. First, let’s use the “Highlighting of matching substructures” feature. You can access this by right-clicking on the structure in the “Structures” view and selecting it from the context menu. This helps us pinpoint areas of the structure that might be incorrect or suspicious. Substructures are color-coded as follows: green substructures are supported by fingerprint evidence, pink substructures contradict fingerprint evidence, yellow substructures have mixed support, and uncolored substructures are not clearly covered by fingerprint evidence.
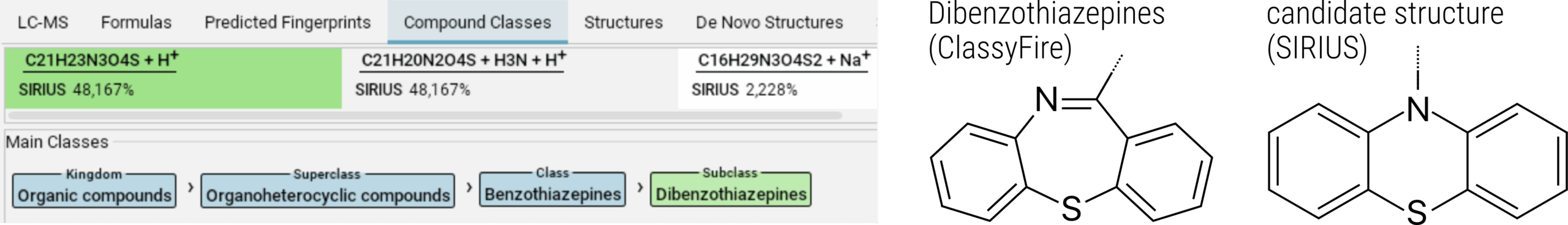
Next, let’s examine the “Compound Classes” view. Our feature is categorized as a dibenzothiazepine, which are derivatives of thiazepine featuring two benzene rings. Clicking on the compound class will open the ClassyFire website, giving us a visual representation of such structures. While our best candidate structure is tricyclic, it lacks the central thiazepine ring. This immediately signals a discrepancy.
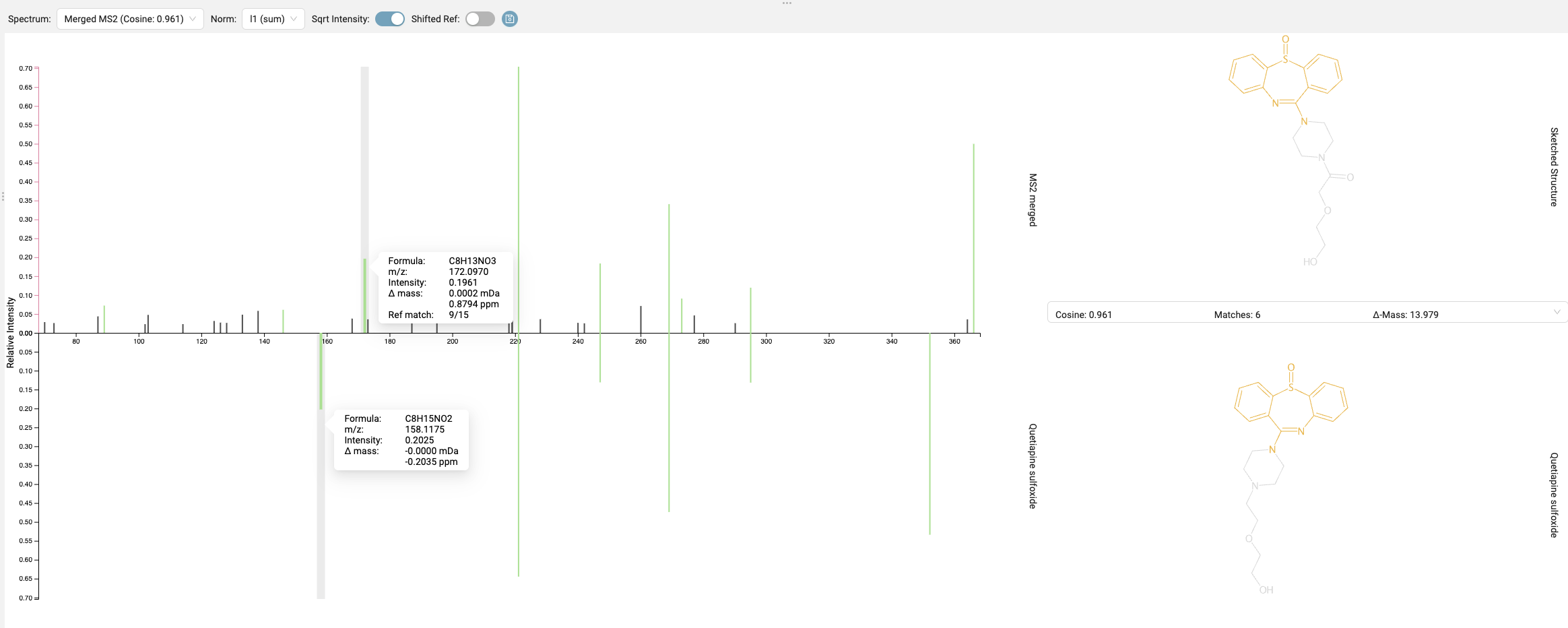
Last, we look at the “Substructure Annotation” results. The best candidate structure might not fully explain some of the fragment peaks in the mass spectrum. By clicking the “sqrt intensity” button, you can reveal smaller peaks. Peaks highlighted in blue are explained by a molecular formula in the fragmentation tree but not by a substructure of the candidate using combinatorial fragmentation. These unexplained peaks are crucial indicators that our current candidate might be inaccurate.
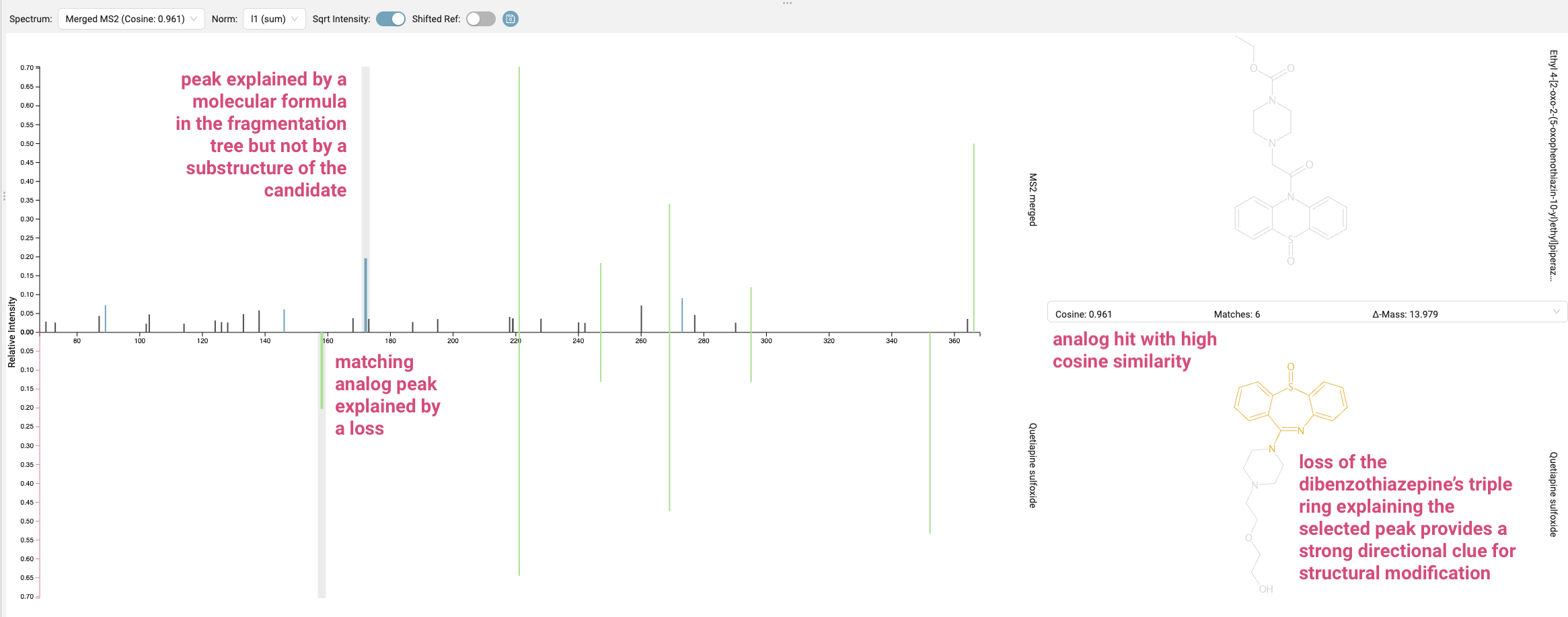
Even if you don’t find exact identity hits from a spectral library search, analog hits can be incredibly valuable for explaining fragments. Our spectrum shows high cosine similarity to Quetiapine sulfoxide, a known dibenzothiazepine. By selecting one of the previously unexplained peaks in our query spectrum and matching it in the analog spectrum, we observe that it’s explained by the loss of the dibenzothiazepine’s triple ring. This provides a strong directional clue for structural modification.
Introducing the Structure Sketcher
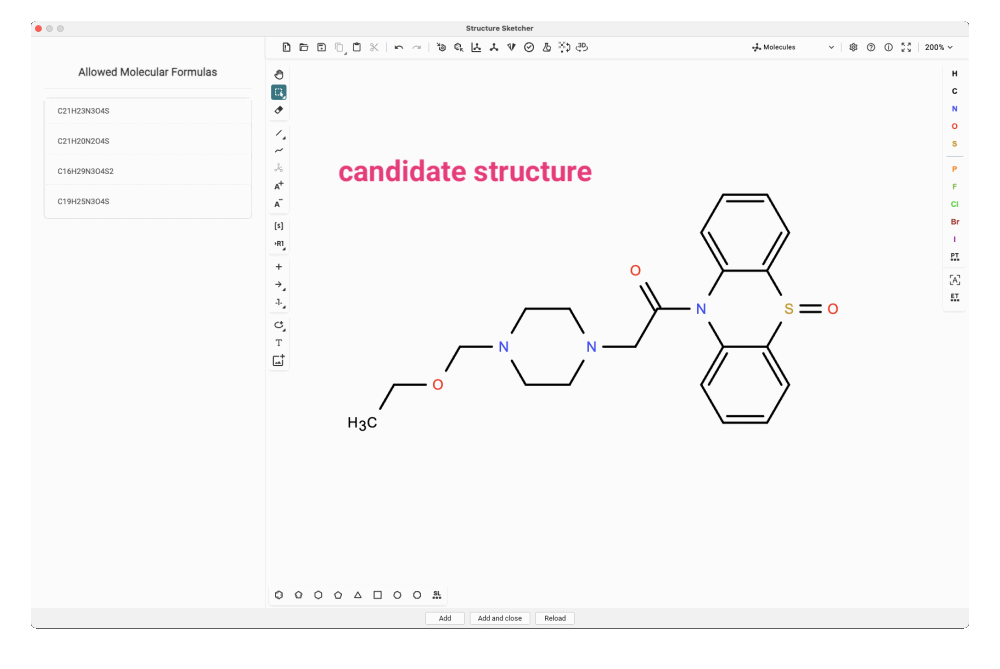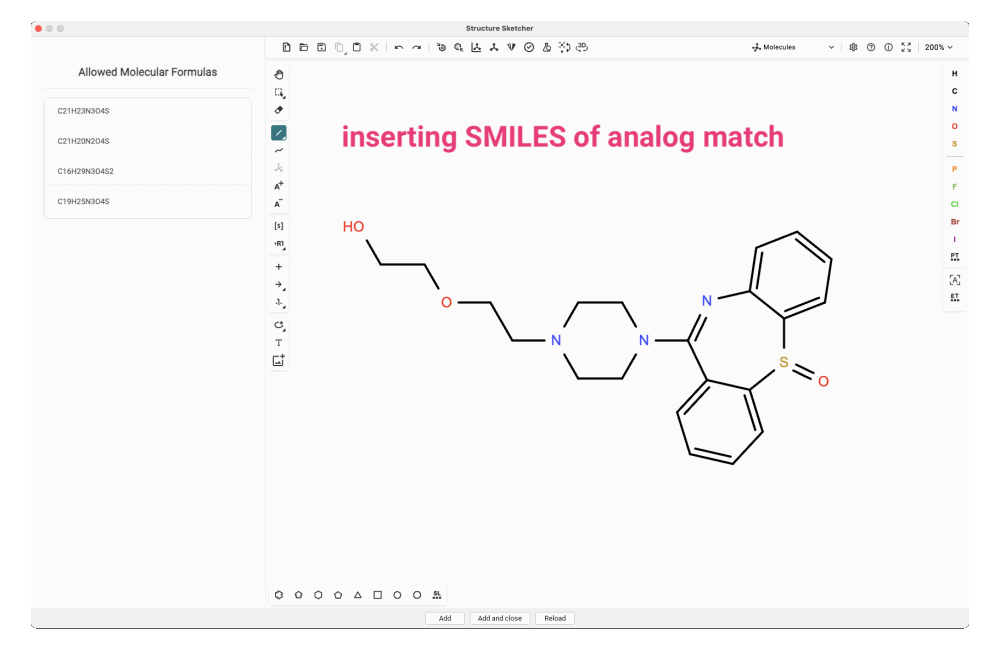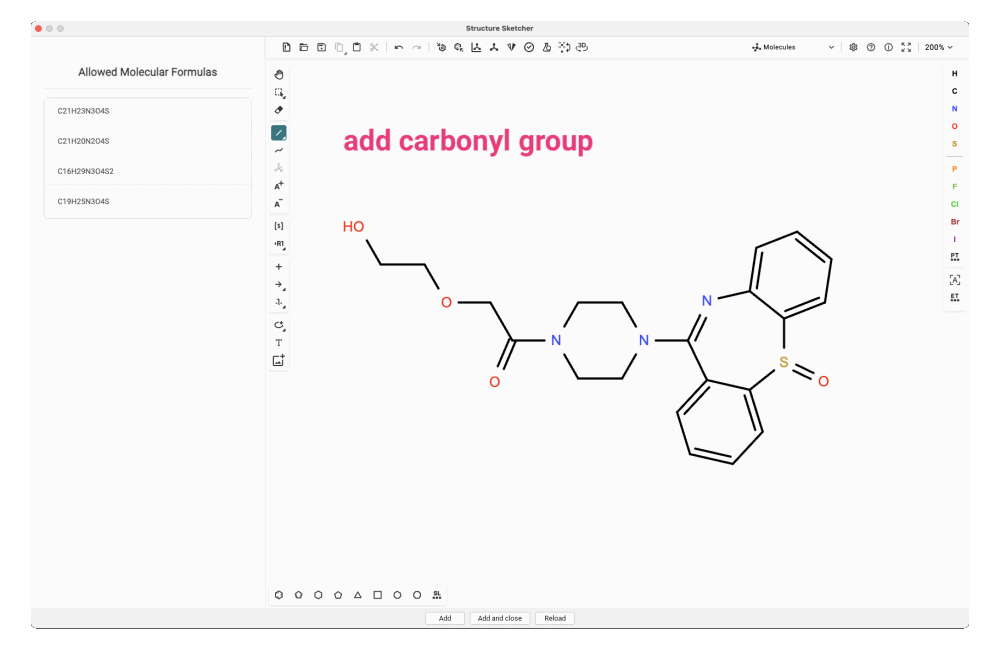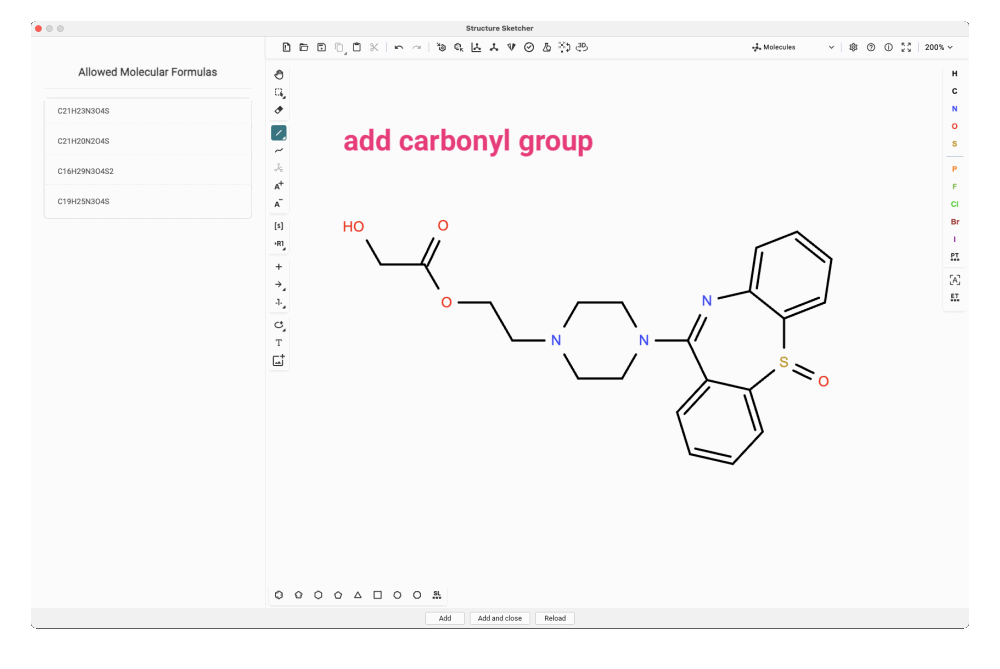
When multiple validation methods (like compound class prediction and analog hits) converge, pointing towards a specific structural motif, it’s time to consider modifying your candidate structure accordingly. In the “Structures” view in SIRIUS, you can right-click on the candidate and select “Modify structure” from the context menu to launch the Structure Sketcher in a separate window. The Structure Sketcher is designed for manually modifying candidate structures and integrating these new structures into your list of candidates.
The sketching area is based on Ketcher, an open-source web-based chemical structure editor. This is where you’ll draw and modify chemical structures. On the left, you’ll find a list of allowed molecular formulas, which includes all molecular formula candidates for which a fingerprint has been predicted. Once your modifications are complete, click Add to include the new structure in your candidate list.
When modifying a candidate structure, it is crucial to ensure that the altered structure conforms to one of the allowed molecular formulas. Failure to do so will result in an error message indicating that no fingerprint was predicted for the generated molecular formula.
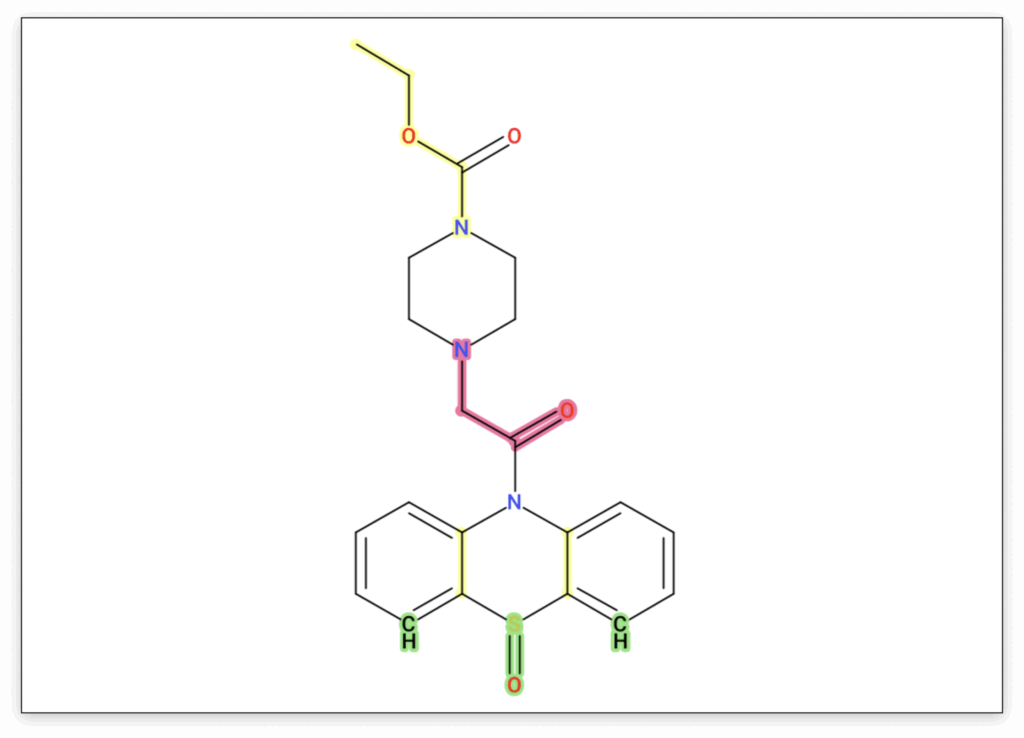
For our feature, we craft the dibenzothiazepine triple ring into our candidate structure. In this particular case, it might be even simpler to copy and paste the SMILES string of the analog match and modify that structure. To match one of the allowed molecular formulas, we need to add the missing carbonyl group. We try adding it at different positions in the side chain and add the new structures to the candidate list.
Evaluating Your Modified Candidates
After closing the Structure Sketcher, we switch to the “De Novo Structures” view to observe our newly added candidates. SIRIUS offers various filters in this view to help you manage and review the updated list. For instance, you can toggle the display of database hits or use the database filter to show only your manually sketched candidates or all structure candidates. You might find that your new candidate(s) score worse than the original. Use the highlighting again to reveal suboptimal placement of substructures.
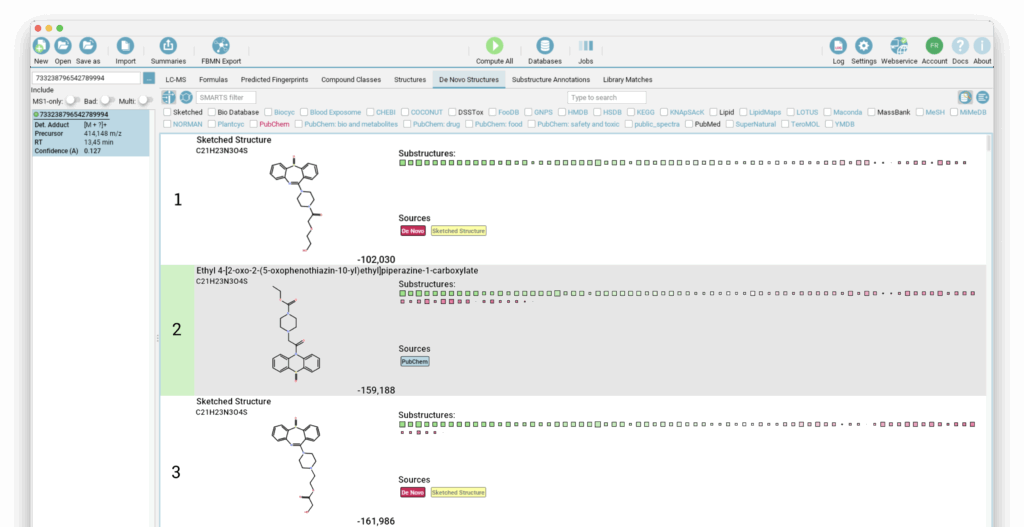
In our example, our modified structure achieves a much better score than the original candidate, and the highlighting clearly suggests that our structural rearrangement was effective. In the Substructure Annotations view, we can further verify whether our new candidate provides better explanations for the observed peaks. Here we can see that a previously unexplained peak can now be clearly attributed to the dibenzothiazepine loss, and other peaks are explained by the newly added side chain.
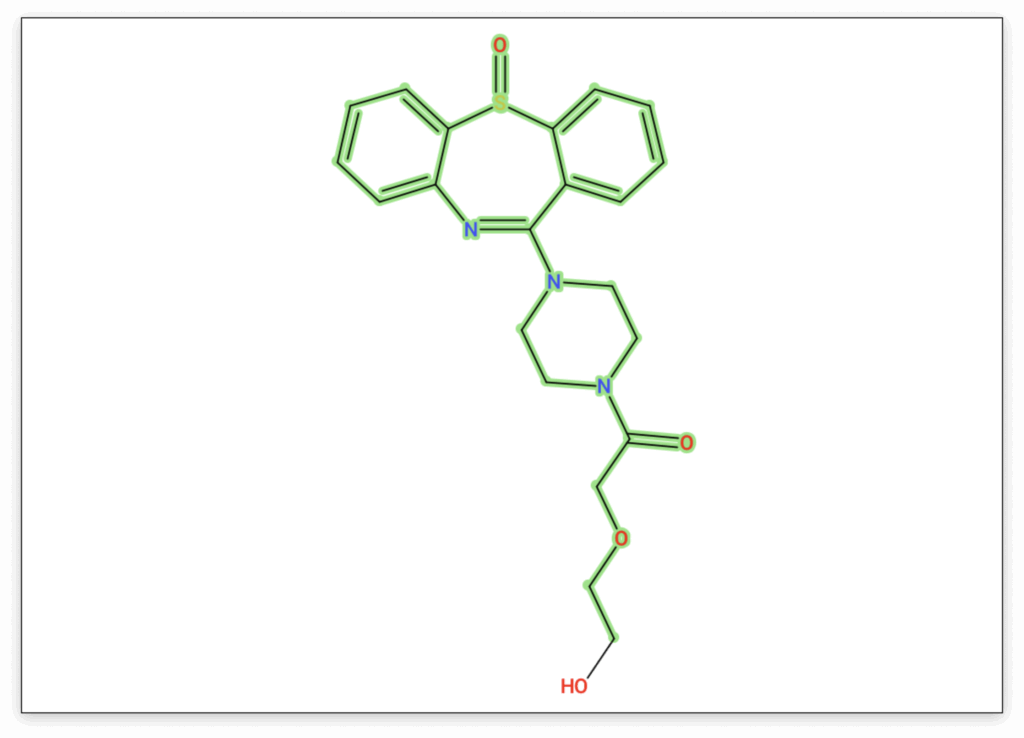
By effectively utilizing the Structure Sketcher and integrating insights from various SIRIUS views, you can manually refine your candidate structures, leading to more accurate annotations and a deeper, more robust understanding of your mass spectrometry data.
Ready to put your chemical intuition to the test with SIRIUS?